Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor
Division of Agricultural Genetics • Kakutani Group
Yu Fu, Akira Kawabe, Mathilde Etcheverry, Tasuku Ito, Atsushi Toyoda, Asao Fujiyama, Vincent Colot, Yoshiaki Tarutani, Tetsuji Kakutani
EMBO Journal, advance online publication 30 July 2013; doi:10.1038/emboj.2013.169
Transposable elements (TEs) have a major impact on genome evolution, but they are potentially deleterious, and most of them are silenced by epigenetic mechanisms such as DNA methylation. Here we report the characterization of a TE encoding an activity to counteract epigenetic silencing by the host. In Arabidopsis thaliana, we identified a mobile copy of the Mutator-like element (MULE) with degenerated terminal inverted repeats (TIRs). This TE, named Hiun (Hi), is silent in wild type plants, but it transposes when DNA methylation is abolished. When a Hi transgene was introduced into the wild type background, it induced excision of the endogenous Hi copy, suggesting that Hi is the autonomously mobile copy. In addition, the transgene induced loss of DNA methylation and transcriptional activation of the endogenous Hi. Most importantly, the trans-activation of Hi depends on a Hi-encoded protein different from the conserved transposase. Proteins related to this anti-silencing factor, which we named VANC, are widespread in the non-TIR MULEs and may have contributed to the recent success of these TEs in natural Arabidopsis populations.
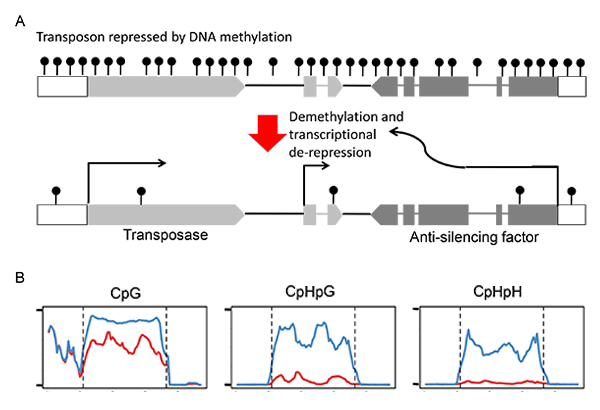
(A) Structure of the plant transposon Hiun (Hi). This transposon encodes for the transposase and the anti-silencing factor.
The antisilencing factor induces demethylation and transcriptional de-repression.
(B) DNA methylation of endogenous Hi compared between non-transgenic plant (blue) and transgenic plant expressing Hi transgene (red).
The transgene induced demethylaton of endogenous copy, especially at non-CpG sites.
Wnt/Dkk negative feedback regulates sensory organ size in zebrafish
Division of Molecular and Developmental Biology • Kawakami Group
H. Wada, A. Ghysen, K. Asakawa, G. Abe, T. Ishitani, K. Kawakami.
Current Biology, 23 (16), p1559–1565, DOI: 10.1016/j.cub.2013.06.035
Correct organ size must involve a balance between promotion and inhibition of cell proliferation. A mathematical model has been proposed, where an organ is assumed to produce its own growth activator, as well as a growth inhibitor. But, there is yet no molecular evidence to support this model. The mechanosensory organs of the fish lateral line system (neuromasts) are composed of a core of sensory hair cells surrounded by non-sensory support cells. Sensory cells are constantly replaced, and are regenerated by surrounding non-sensory cell, while each organ retains the same size throughout life. Moreover, neuromasts also bud off new neuromasts, which stop growing when they reach the same size. In this study, we show that the size of neuromasts is controlled by a balance between growth-promoting Wnt signaling activity in proliferation-competent cells, and Wnt-inhibiting Dkk activity produced by differentiated sensory cells. This negative feedback loop from Dkk (secreted by differentiated cells) on Wnt-dependent cell proliferation (in surrounding cells) also acts during regeneration to achieve size constancy. This study establishes Wnt/Dkk as a novel mechanism to determine the final size of an organ.
This study was funded by the PRESTO of the Japan Science and Technology Agency.

A) Schematic drawing of a neuromast.
(B) Schematic representation of Wnt signaling and of its inhibition by Dkk signaling.
(C) Wnt reporter activity (green) gradually subsides as hair cells (red) are formed.
(D) Dkk2 expression coincides with neuromast maturation.
Detailed analysis of a complex copy number variation region in mouse genome
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Umemori J, Mori A, Ichiyanagi K, Uno T, and Koide T.
BMC Genomics14: 455, 2013.
Copy number variation (CNV) of genomic segments is a common phenomenon that affects more than 10% of the human and mouse genomes, and is an important source of diversity in genomic structure. CNVs are frequently found in clusters called CNV regions (CNVRs), which are strongly associated with large segmental duplications (large SDs). However, the composition of these complex repetitive structures remains unclear. In the present study, we established new method for analyzing on complex repetitive structures of CNVRs by collaborating with National Institute of Informatics.
At first, we conducted self-comparative-plot analysis of all mouse chromosomes using the high-speed and large-scale-homology search algorithm, Similarity/Homology Efficient Analyze Procedure (SHEAP) developed by Dr. Takeaki Uno in National Institute of Informatics. By using this method, large SDs were visualized as unique tartan-checked patterns with complex arrangement of diagonal split lines (Figure 1). We focused on one SD on chromosome 13 (SD13M), which is one of the causative regions for genetic incompatibility (papers in preparation), and applied blast-based Systematic analysis of HErPlot to Extract Regional Distinction (SHEPHERD), a stepwise ab initio method, to extract core elements of repetitive sequences in SD13M (Figure 2). Then, comparative genome hybridization array analysis was empirically conducted on MSM, BLG2 (strains derived from wild mice in Mishima in Japan, and Toshevo in Bulgaria, respectively), and C57BL/6J (an experimental strain). This analysis showed that core elements are categorized to ones with CNVs and the others with constant copy number among strains, which have distinctive characters and divergences. The present study seems to be helpful for elucidating evolutional processes and functions of the CNVRs (Fig. 3).
This study was funded by the “Cultivation of integrated project” of Transdisciplinary Research Integration Center in Research Organization of Informatio and Systems (Japan).
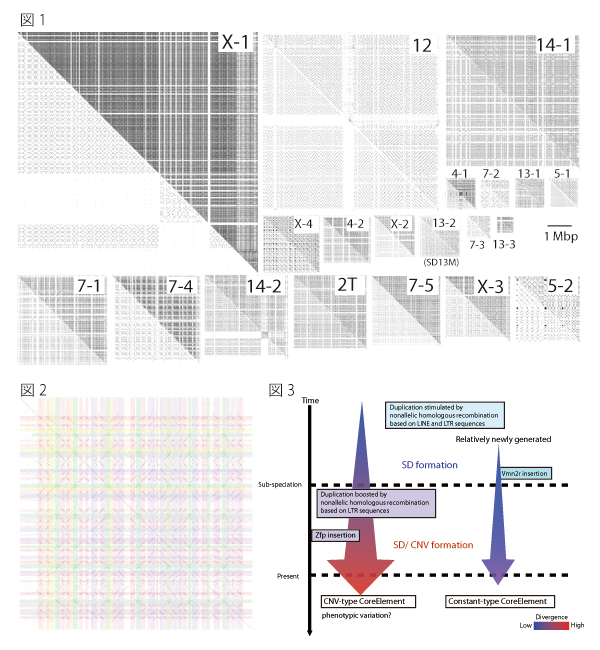
Fig.1. Tartan-checked structure of SDs visualized using SHEAP.
The lower left triangle of each panel shows a self-plot of the sequence after known repeat sequences have been masked using RepeatMasker. Each of the upper right triangles shows a self-plot of the intact sequence. Diagonal lines aligned in the same column or row represent repetitive sequences.
Fig.2. Extraction of repeat units from the self-plot of the large SD
Diagonal lines were extracted from a self-comparative-plot of SD13M that consisted of a dot-plot matrix. Then we selected one sequence and removed the other sequences represented by diagonal lines that were located in the same column or in the same row.
Fig.3 Model for the formation of SDs and CNVs
The average divergence of CNV-type core elements was greater than that of the constant type, and the CNV-type core elements contained significantly larger proportions of long terminal repeat (LTR) types of retrotransposon than the constant-type core elements. These results suggest that constant-type core elements emerged more recently than CNV-type core elements, and that retrotransposition of LTRs promotes nonallelic homologous recombination and caused CNV in SD13M.
Concerted interaction between origin recognition complex (ORC), nucleosomes, and replication origin DNA ensures stable ORC–origin binding
Division of Microbial Genetics • Araki Group
Kohji Hizume, Masaru Yagura, and Hiroyuki Araki
Genes to Cells , 24 JUN 2013 DOI: 10.1111/gtc.12073. [Epub ahead of print]
Chromosomal replication origins, where DNA replication is initiated, are determined in eukaryotic cells by specific binding of a six-subunit origin recognition complex (ORC). Many biochemical analyses have revealed the detailed properties of the ORC–DNA interaction. However, because of the lack of in vitro analysis, the molecular architecture of the ORC–chromatin interaction is unclear. Recently, mainly from in vivo analyses, a role of chromatin in the ORC–origin interaction has been reported, including the existence of a specific pattern of nucleosome positioning around origins and of a specific interaction between chromatin—or core histones—and Orc1, a subunit of ORC. Therefore, to understand how ORC establishes its interaction with origin in vivo, it is essential to know the molecular mechanisms of the ORC–chromatin interaction. Here, we show that ORC purified from yeast binds more stably to origin-containing reconstituted chromatin than to naked DNA, and forms a nucleosome-free region at origins. Molecular imaging using atomic force microscopy (AFM) reveals that ORC associates with the adjacent nucleosomes and forms a larger complex. Moreover, stable binding of ORC to chromatin requires linker DNA. Thus, ORC establishes its interaction with origin by binding to both nucleosome-free origin DNA and neighboring nucleosomes.
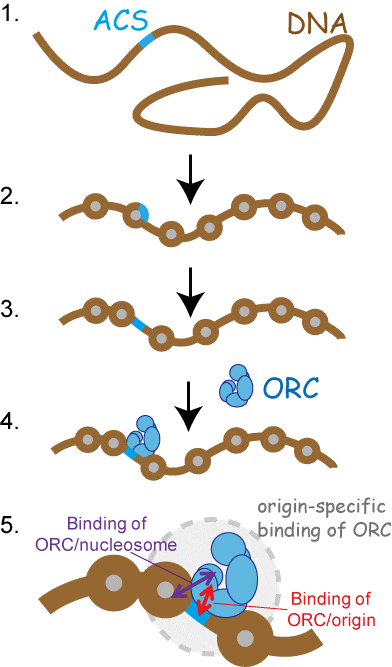
(1) DNA fragment containing origin-specific DNA motif (ACS).
(2) By chromatin reconstitution, nucleosome is formed on ACS.
(3) By the addition of ORC, ACS becomes nucleosome-free-region.
(4) ORC interacted to nucleosome-free ACS.
(5) ORC establishes its interaction with origin by binding to both nucleosome-free ACS and neighboring nucleosomes.
Regulation of nuclear envelope dynamics that is necessary for the progression of semi-open mitosis
Microbial Genetics Laboratory • Niki Group
Keita Aoki, Yuh Shiwa, Hiraku Takada, Hirofumi Yoshikawa and Hironori Niki
Genes to Cells 21 JUN 2013 DOI: 10.1111/gtc.12072. [Epub ahead of print]
Three types of mitosis, which are open, closed, or semi-open mitosis, function in eukaryotic cells, respectively. The open mitosis involves breakage of the nuclear envelope before nuclear division, whereas the closed mitosis proceeds with an intact nuclear envelope. To understand the mechanism and significance of three types of mitotic division in eukaryotes, we investigated the process of semi-open mitosis, in which the nuclear envelope is only partially broken, in the fission yeast Schizosaccharomyces japonicus. In anaphase-promoting complex/cyclosome (APC/C) mutants of Sz. japonicus, the nuclear envelope remained relatively intact during anaphase, resulting in impaired semi-open mitosis. As a suppressor of apc2 mutant, a mutation of Oar2 which was a 3-oxoacyl-[acyl-carrier-protein] reductase was obtained. The level of the Oar2, which had two destruction-box motifs recognized by APC/C, was increased in APC/C mutants. Furthermore, the defective semi-open mitosis observed in an apc2 mutant was restored by mutated oar2+. Based on these findings, we propose that APC/C regulates the dynamics of the nuclear envelope through degradation of Oar2 dependent on APC/C during the metaphase-to-anaphase transition of semi-open mitosis in Sz. japonicus.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
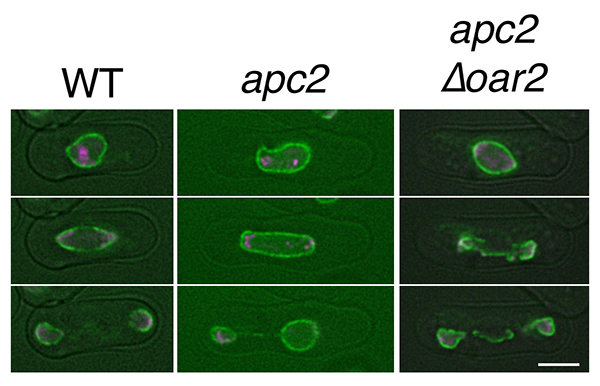
Observations of nuclear envelope dynamics in WT, apc2 mutant, and apc2∆oar2 mutant. The semi-open mitosis appeared in WT was defective in apc2 mutant, but partially restored in apc2∆oar2 mutant. Green: nuclear envelope marker (Cut11-GFP), magenta: chromosomal marker (H2A-mCherry). Scale bar: 5µm.















