Chromatin dynamics in human transformed cells
Maeshima Group / Genome Dynamics Laboratory
Single-nucleosome imaging uncovers biphasic chromatin dynamics in inducible human transformed cells
Aoi Otsuka, Masa A. Shimazoe, Shigeaki Watanabe, Katsuhiko Minami, Sachiko Tamura, Tohru Kiyono, Fumitaka Takeshita, and Kazuhiro Maeshima* (*Corresponding author)
Cell Structure and Function (2025) Advance online publication DOI:10.1247/csf.25147
In eukaryotic cells, genomic DNA folds into nucleosomes to form dynamic domains encompassing euchromatin and heterochromatin. Although many cancer-associated changes in chromatin state and higher-order structure have been reported, how chromatin behavior evolves over time during carcinogenesis has remained unclear.
SOKENDAI students Aoi Otsuka (SOKENDAI Special Researcher) and Masa A. Shimazoe (JSPS DC1), postdoctoral researcher Katsuhiko Minami, technical staff member Sachiko Tamura, and Professor Kazuhiro Maeshima of the Genome Dynamics Laboratory, in collaboration with Visiting Researcher Dr. Tohru Kiyono (also a Visiting Researcher at the Sasaki Institute (Sasaki Foundation)), Project Researcher Dr. Shigeaki Watanabe, and Division Chief Dr. Fumitaka Takeshita at the National Cancer Center, established “EMR” human epithelial cells that inducibly express the oncogenes HPV16 E6/E7, MYC, and KRAS upon doxycycline treatment. Upon induction, EMR cells exhibited cancer-like traits—accelerated proliferation, loss of contact inhibition, soft-agar growth, and tumor formation in nude mice. Live-cell single-nucleosome imaging revealed a biphasic pattern in chromatin dynamics: no change at days 1–3, a transient increase at days 5–7, and a return to baseline by week 4. During this window of increased mobility, histone H3/H4 acetylation and transcription were elevated. Together, these results suggest that oncogene induction causes a transient chromatin “loosening” accompanied by widespread acetylation and transcriptional activation, followed by restabilization of chromatin dynamics even while oncogene expression and tumor growth persist.
This work demonstrates at the single-nucleosome level that the physical behavior of chromatin is reorganized over time during transformation, and that chromatin dynamics can serve as a physical readout of the cancer stage and cellular adaptation.
Funding: JSPS and MEXT KAKENHI (JP23K17398, JP24H00061, JP23KJ0998, JP24KJ1161), JST SPRING JPMJSP2104, and the Takeda Science Foundation.
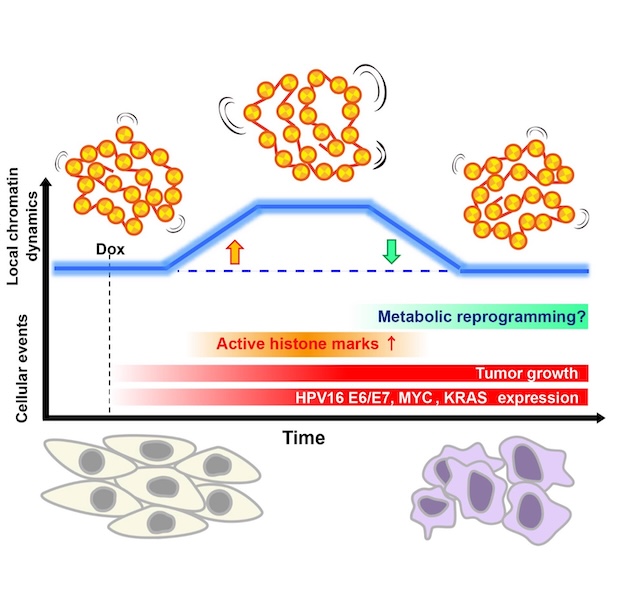
Figure: Model summarizing the biphasic change in local chromatin dynamics during doxycycline-induced transformation of EMR cells. The blue curve denotes a transient rise in local chromatin mobility after induction (orange arrow) followed by a return to baseline (green arrow). The increase coincides with elevated active histone marks (H3/ H4 acetylation) and transcription; dynamics subsequently restabilize while oncogene expression and tumor growth persist. Metabolic reprogramming may contribute to this process.















