Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Discovery of the Oldest Sex Chromosomes among Vertebrates
—Unique Sex Determination mechanisms in Sharks and Rays —
Press release
Sharks and rays have the oldest vertebrate sex chromosome with unique sex determination mechanisms
Taiki Niwa, Yoshinobu Uno, Yuta Ohishi, Mitsutaka Kadota, Naotaka Aburatani, Itsuki Kiyatake, Daiki Katooka, Michikazu Yorozu, Nobutaka Tsuzuki, Atsushi Toyoda, Wataru Takagi, Masaru Nakamura, Shigehiro Kuraku*
* Corresponding Author
Proceedings of the National Academy of Sciences USA (2025) DOI:10.1073/pnas.2513676122
Summary
Many animals have sexes, but the mechanisms that determine them are remarkably diverse. In vertebrates, sex can be influenced by genetic factors or environmental cues such as temperature. However, how these diverse systems evolved remains one of major unanswered questions in biology.
Cartilaginous fishes, such as sharks and rays, represent one group deeply isolated from most of vertebrates, providing a unique window to vertebrate evolution. Despite their evolutionary importance, little has been known about how sex is determined in these groups. The research team, including graduate student Taiki Niwa and Professor Shigehiro Kuraku both in Molecular Life History Laboratory and Atsushi Toyoda in Comparative Genomics Laboratory at NIG, investigated genome sequences of various cartilaginous fish species. Their work, published in Proc. Natl. Acad. Sci. USA, revealed that sharks and rays share a common set of genes on their X chromosomes, indicating that this sex chromosome system is estimated to have maintained for nearly 300 million years, which aligned with the massive loss of their gene content of Y chromosomes.
This study also found that these X chromosomes lack dosage compensation, which is used in mammals and other vertebrates to balance gene expression from sex chromosomes between sexes. This absence of dosage compensation suggests that gene dosage play a central role in sex determination in sharks and rays. These findings highlight a distinct evolutionary mode of their sex determination, and provide new insight into the evolution of sex determination mechanisms.

Research Background
Sex chromosomes are one of mechanisms that determine sex in vertebrates. In humans, individuals typically inherit 23 pairs of chromosomes from their parents. One of them is composed of chromosomes with different sizes, namely X and Y chromosomes, with the presence of the Y chromosome triggering male development. In contrast, most chromosomes (called autosomes) are similar between the two copies in their sizes, shedding light on the distinct evolutionary mode of sex chromosomes.
Among vertebrates, mammals and birds have ancient sex chromosomes that emerged over 100 million years ago, and their X and Y chromosomes differ significantly in size and gene content. Many ray-finned fishes, however, have much younger sex chromosomes that still resemble each other closely. From the accumulation of these observations, researchers have inferred the typical evolutionary path of sex chromosomes that they evolve from similar-looking pairs that diverge over time, with one gradually losing genes. Yet, many aspects of this process remain unknown.
Although cytogenetic studies by microscopic observations had suggested that some cartilaginous fishes possess an XX/XY sex chromosome system, the underlying molecular mechanism and evolutionary history had not been understood (Fig. 1).

Figure1: An overview of the sex determination systems found across major vertebrate lineages, based on a broad accumulation of previous studies (refer to citations in the original article).
Key Findings
First, the team generated high-quality whole genome sequences for two shark species: the cloudy catshark (Scyliorhinus torazame) and the brownbanded bamboo shark (Chiloscyllium punctatum). These genome sequences represent a major improvement over a version previously produced by the team, which has continuously pursued genome-wide data collection for sharks and rays under the dedicated consortium Squalomix.
By comparing the genomes of these two species with those of other cartilaginous fishes whose genome data had recently become available, the team identified and compared the sequences of sex chromosomes. They found that a wide range of shark and ray species share a conserved set of genes on their X chromosomes (Fig. 2). This suggests that these sex chromosomes have remained unchanged since the last common ancestor of these species, which lived about 300 million years ago. This origin predates those of the formerly oldest sex chromosomes in mammals and birds, which are estimated to have originate 100–200 million years ago.

Figure2: A comparison of X chromosome sequences among cartilaginous fishes. Species whose whole-genome sequences were newly assembled by the research team are shown in bold. On the right, dark horizontal bars represent chromosomes, labeled with chromosome IDs (numbers or letters), and bands between them indicate conserved gene arrangements. Similarity in gene content was observed across the X chromosomes (highlighted in dark orange) in sharks and rays. In the red stingray, two X chromosomes (X1 and X2) were identified. Portions of these chromosomes correspond to autosomes (gray) in other species, suggesting a fusion between sex chromosomes and autosomes. Chimaeras, which are phylogenetically distant from sharks and rays, showed no evidence of X chromosomes homologous to those found in sharks and rays.
In addition, the team successfully isolated and sequenced the Y chromosome of the bamboo shark by using a previously established cell culture system. The Y chromosome was found to have significantly fewer genes than the X chromosome, confirming the degradation that typically occurs over long evolutionary timeframes.
Thanks to the availability of high-quality genome sequences and embryos from aquarium-reared sharks, the team was able to examine the developmental timing and structure of the gonads using histological methods. They determined the stage at which male and female gonads begin to differentiate. Using RNA-seq analysis, they also assessed gene expression in male and female gonads during this critical period. Their findings revealed that genes on the X chromosome are expressed in proportion to their copy number—females, with two X chromosomes, express these genes at higher levels than males, who have only one X (Fig. 3). This suggests the absence of a dosage compensation mechanism like that seen in mammals, where one of the two X chromosomes in females is largely inactivated. Instead, the expression difference in X-linked genes appears to contribute directly to sex determination.
Notably, none of the known sex-determining genes identified in other vertebrates were found on these shark and ray sex chromosomes, indicating that these species may rely on a novel mechanism of sex determination. The long-standing X chromosome and its use in sex determination through gene dosage may reflect a unique evolutionary strategy among sharks and rays.

Figure3: Gene expression ratios between sexes in the developing gonads. The photograph shows a bamboo shark embryo removed from its egg case at the stage when sex is determined. Oviparous cartilaginous fishes spend several months developing within an egg capsule after being laid. The graph presents the distribution of gene-by-gene expression ratios between sexes. In mice, a dosage compensation mechanism inactivates one of the two X chromosomes in females, resulting in similar expression levels between sexes for both autosomal and X-linked genes. In contrast, in the bamboo shark, the majority of 163 expressed X-linked genes were more highly expressed in females, indicating a lack of dosage compensation. The mouse data are based on previously published RNA-seq results from adult liver tissue.
Future Outlook
Sex determination systems vary widely across the animal kingdom, and how they evolve is still largely mysterious. Some have speculated that the human Y chromosome may eventually disappear, but this study shows a counterexample that Y chromosomes can persist over hundreds of millions of years.
By exploring a broader range of organisms like sharks and rays, scientists can better understand the diversity and evolution of sex chromosomes, which may also provide insight into the past and future of our sex.
Masa A. Shimazoe won the “Poster prize: 1st Place”
Masa A. Shimazoe, D4 student and JSPS Special Researcher in Genome Dynamics Laboratory(Maeshima Laboratory), received the “Poster Prize: 1st Place” at the “EMBL Conference: Gene regulation: one molecule at a time” held in EMBL Heidelberg, Germany, on July 15th – 18th. In addition, he also received the “EMBL Advanced Training Centre Corporate Partnership Programme fellowship”.
SOKENDAI Student Dispatch Program supported this trip.
▶ Awarded presentation title: Linker histone H1 functions as a liquid-like glue to organize chromatin in living human cells
▶ EMBL Conference: Gene regulation: one molecule at a time

Masa A. Shimazoe
Samal Tazhibayeva won the “Poster Award”

Samal Tazhibayeva (D4, Department of Genetics, SOKENDAI) in Multicellular Organization Laboratory received the Poster Award at Joint Meeting of JSCB & JSDB Held in Nagoya on July 16th-18th.
・ Awarded presentation title: CWN-2/Wnt localization in C. elegans challenges gradient-dependent
polarity regulation by Wnt
Summer Holiday (Aug. 14-15)
NIG will be closed from August 14 to August 15, 2025 for summer holiday.
Thank you for your understanding and cooperation.
Samal Tazhibayeva won the “Poster Award (Honorable Mention)”
Samal Tazhibayeva (D4, Department of Genetics, SOKENDAI) in Multicellular Organization Laboratory received the Poster Award (Honorable Mention) at the 25th International Worm Meeting held in UC Davis USA on June 28th-July 2nd 2025. She also received the grand prize in Art Show during the meeting by her painting.
・ Awarded presentation title: CWN-2/Wnt localization on seam cells challenges gradient-dependent polarity regulation by Wnt
A new technology that enables degradation of target proteins without fusing a degron tag.
Kanemaki Group / Molecular Cell Engineering Laboratory
Sawa Group / Multicellular Organization Laboratory
A Single-Chain Antibody-Based AID2 System for Conditional Degradation of GFP-Tagged and Untagged Proteins
Moutushi Islam, Takefumi Negishi, Naomi Kitamoto, Yuki Hatoyama, Kanae Gamo, Ken-ichiro Hayashi, and Masato T. Kanemaki*
*corresponding author
Journal of Cell Science (2025) jcs.263961 DOI:10.1242/jcs.263961
By inducing rapid degradation of a target protein in living cells, it becomes possible to observe the effects caused by the loss of that protein’s function. The auxin-inducible degron (AID) technology developed by the Kanemaki Laboratory has been widely used for functional studies of various proteins. The latest improved version, AID2, allows precise and efficient control of target protein degradation. However, AID2 requires genome editing to fuse the degron tag to the target protein, which has limited its application to non-model organisms and clinical settings. To overcome this limitation, the Kanemaki Laboratory successfully developed the single-chain antibody AID2 (scAb-AID2) system, which enables degradation without the need to tag the target protein by utilizing single-chain antibodies.
As a proof-of-concept experiment, they employed a GFP nanobody, a single-chain antibody that recognizes GFP. An adapter consisting of the GFP nanobody fused to a degron tag was introduced along with the ubiquitin ligase OsTIR1(F74G) into human cultured cells and C. elegans expressing various GFP-tagged proteins. Upon addition of the degradation-inducing ligand 5-Ph-IAA, these GFP-fusion proteins were successfully degraded.
Furthermore, they designed adapters fused with degron tags using single-chain antibodies targeting the tumor suppressor p53 and the growth-promoting factors H/K-RAS. These adapters and OsTIR1(F74G) were introduced into human cultured cells. Treatment of these cells with 5-Ph-IAA induced degradation of p53 and H/K-RAS. Notably, degradation of p53 increased the cells’ sensitivity to a replication stress-inducing agent, while tumors formed from cells in which H/K-RAS was degraded showed suppressed tumor formation.
The scAb-AID2 system overcomes the limitation of degron tagging required in the conventional AID2 system and opens new avenues for both basic life science research and clinical applications through targeted protein degradation.
In connection with this paper, the first author, SOKENDAI student Moutushi Islam, was featured in the First Person section of the Journal of Cell Science. In addition, the paper was selected as a Research Highlight in the same journal.
This work was supported by JSPS KAKENHI grant (JP23K05836) to Takefumi Negishi, and JSPS KAKENHI grants (JP21H04719, JP23H04925 and JP25H00979) and JST CREST grant (JPMJCR21E6) to Masato T. Kanemaki.
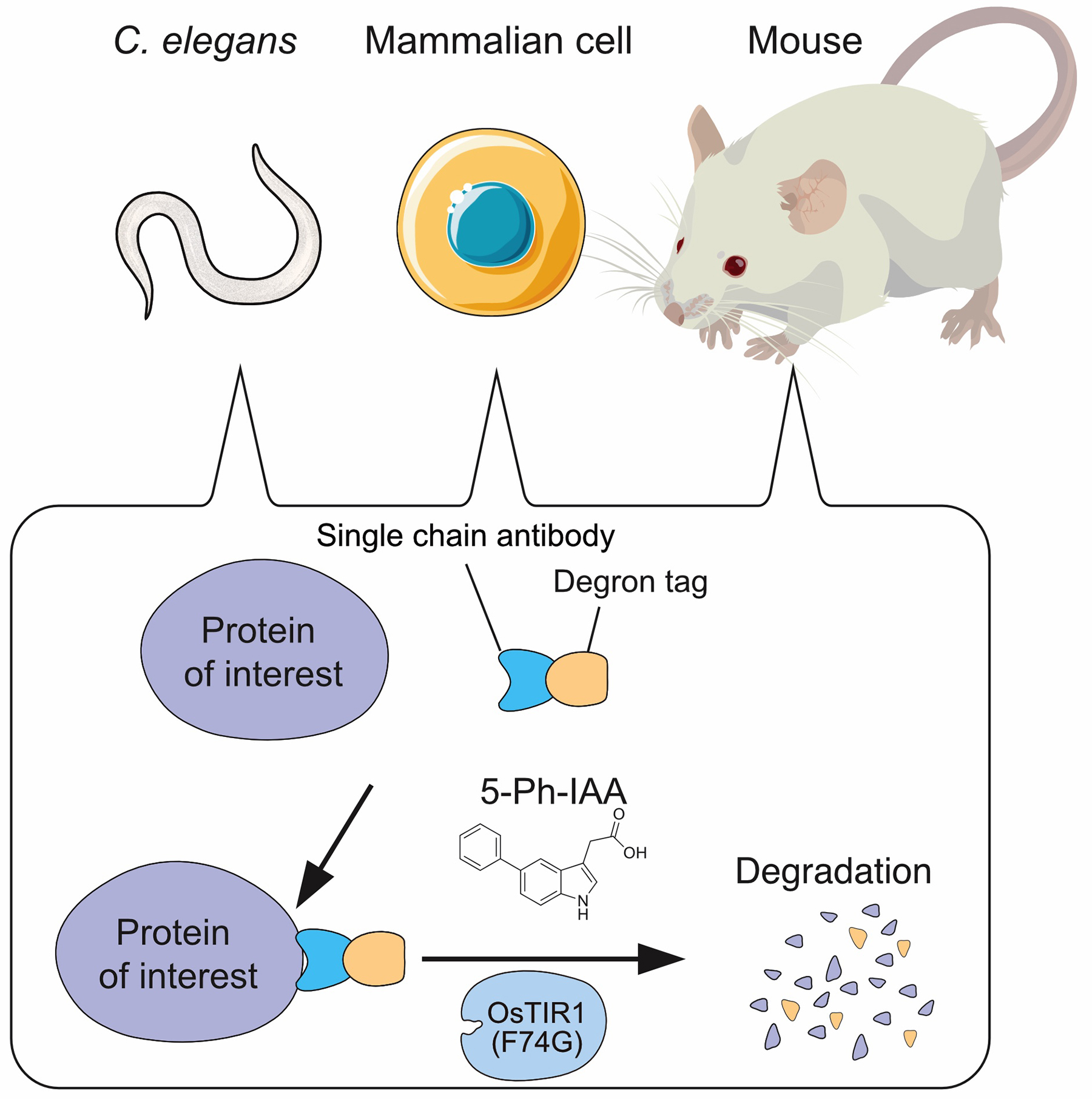
Figure: Single-chain antibody AID2 system enables degradation of untagged proteins in C. elegans, mammalian cells and mouse in a 5-Ph-IAA dependent manner.















