Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Guideline for Additional Application for 2024 NIG-JOINT(Joint Researchi-(A)) (Application was closed)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Guideline for Additional Application for
2024 NIG-JOINT(Joint Researchi-(A))
(Application deadline: noon(12:00pm)
on Friday, May 31st, 2024)
Unveiling the mysteries of cell division in embryos with timelapse photography
Kanemaki Group / Molecular Cell Engineering Laboratory
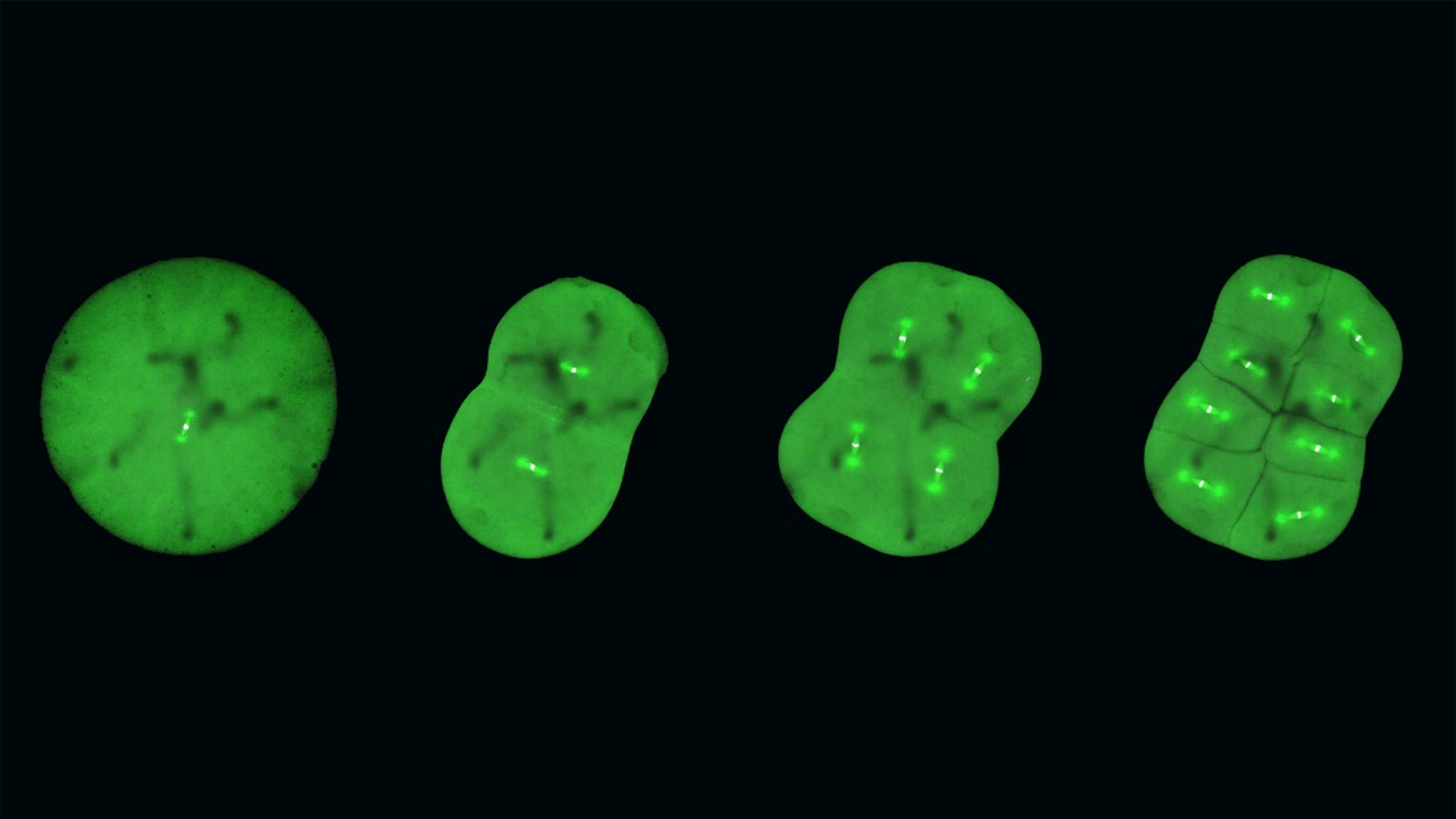
Ran-GTP assembles a specialized spindle structure for accurate chromosome segregation in medaka early embryos
Ai Kiyomitsu, Toshiya Nishimura, Shiang Jyi Hwang, Satoshi Ansai, Masato T. Kanemaki, Minoru Tanaka & Tomomi Kiyomitsu
Nature Communications (2024) 15, 981 DOI:10.1038/s41467-024-45251
With the help of medaka fish, CRISPR and new imaging techniques, researchers have set a new standard for studying cell division at the very earliest stages of life.
The beginning of life is shrouded in mystery. While the intricate dynamics of mitosis is well-studied in the so-called somatic cells – the cells that have a specialized function, like skin and muscle cells – they remain elusive in the first cells of our bodies, the embryonic cells. Embryonic mitosis is notoriously difficult to study in vertebrates, as live functional analyses and -imaging of experimental embryos are technically limited, which makes it hard to track cells during embryogenesis.
However, researchers from the Cell Division Dynamics Unit at the Okinawa Institute of Science and Technology (OIST) have recently published a paper in Nature Communications, together with Professors Toshiya Nishimura from Hokkaido University (previously at Nagoya University), Minoru Tanaka from Nagoya University, Satoshi Ansai from Tohoku University (currently at Kyoto University), and Masato T. Kanemaki from the National Institute of Genetics. The study takes the first major steps towards answering questions about embryonic mitosis, thanks to a combination of novel imaging techniques, CRISPR/Cas9 genome editing technology, a modern protein-knockdown system, and medaka, or Japanese rice fish (Oryzias latipes). The timelapses that they have produced help answer fundamental questions about the intricate process of equally dividing chromosomes during embryonic mitosis, and simultaneously chart the next frontier of scientific exploration. As Professor Tomomi Kiyomitsu, senior author of the study, describes the timelapses: “they are beautiful, both on their own and because they lay a new foundation for elucidating embryonic mitosis.”
Central to the mystery of embryonic mitosis is the crucial step when the chromosomes, which contain all the genetic information of the cell, are aligned and segregated equally into daughter cells. A key player in this process is the mitotic spindle, which is made of microtubules – long protein fibers used for intra-cellular structure and transport – that radiates from opposite poles of the spindle and attaches to the chromosomes in the middle. The spindle captures duplicated chromosomes properly and segregates them equally into the daughter cells during division. There are many factors determining spindle formation, and one of these is the protein Ran-GTP, which plays an essential role in cell division of female reproductive cells, which lack centrosomes – cell organelles responsible for assembling microtubules – but not in small somatic cells, which do have centrosomes. However, it has long been unclear whether Ran-GTP is required for spindle assembly in vertebrate early embryos, which contain centrosomes but have unique features, like a larger cell size.
In contrast to mammalian early embryos, embryonic cells in fish are transparent and develop synchronously in a uniform, single-cell layer sheet, which makes them significantly easier to track. The medaka turned out to be particularly well-suited for the researchers, as these fish tolerate a wide range of temperatures, produce eggs daily, and have a relatively small genome. Being temperature-tolerant means that the medaka embryonic cells could survive at room temperature , making them particularly suited for long, live timelapse photography.
The fact that medaka produce eggs frequently and have a relatively small genome size makes them good candidates for CRISPR/Cas9-mediated genome editing. With this technology, the researchers have created genetically modified, or transgenic, medaka whose embryonic cells literally highlight the dynamics of certain proteins involved in mitosis.
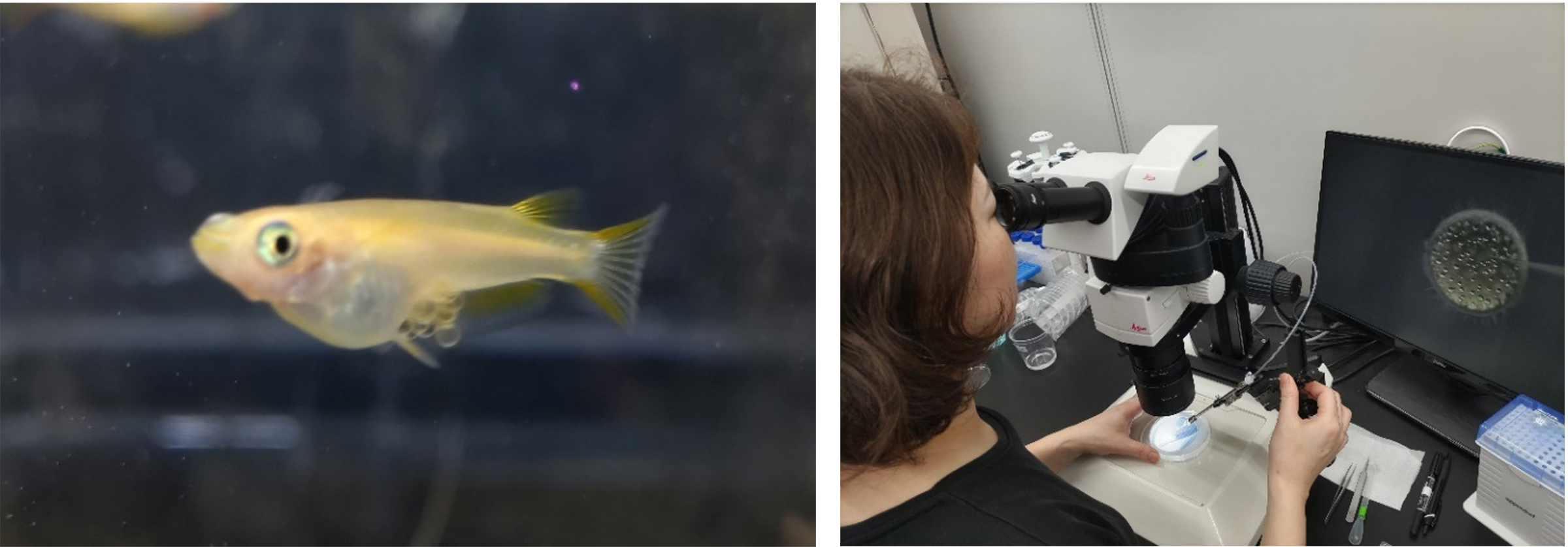
Figure: Left: Egg-carrying medaka fish, used in the study. Right: Dr. Ai Kiyomitsu from the Cell Division Dynamics Unit, first author of the paper, injecting RNA into a medaka embryo as part of the CRISPR/Cas9-mediated genome editing process. Photos: Tomomi Kiyomitsu (OIST).
In studying the timelapses of the developing mitotic spindle in live, transgenic medaka embryos, the researchers discovered that large early embryos assemble unique spindles different from somatic spindles. In addition, Ran-GTP plays a decisive role in spindle formation in early embryonic divisions, but the importance diminishes in later stage embryos. This is possibly because the spindle structure is remodeled as cells get smaller during development, though the exact reason is a subject for future research.
The researchers also discovered that the early embryonic cells do not have a dedicated spindle assembly checkpoint, which characterizes most somatic cells, and which serves to ensure that the chromosomes are properly aligned before segregation. As Professor Kiyomitsu surmises, “the checkpoint is not active, and yet the chromosome segregations are still very accurate. This could be explained by the fact that embryonic cells need to divide very quickly, but it is something that we want to study further.”
While genetically modifying the medaka fish and studying the early embryos have led to new key insights into embryonic mitosis, this is just the beginning for Professor Kiyomitsu and the team. In addition to questions related to the diminishing role of Ran-GTP in later stages and the missing spindle assembly checkpoint, he points to the satisfying symmetry of cell divisions in the timelapses: “The spindle formation is characterized by a high degree of symmetry, as the cells appear to be dividing in the sizes and defined directions, and the spindle is consistently in the center of the cells. How can the spindle orient itself so regularly across the cells, and how is it able to find the center every time?”
Moving beyond the timelapses, the team also hopes to further solidify this new foundation with additional medaka gene-lines to serve as models for research in embryonic cells, and at the same time optimize the genome editing process. Eventually, the team wants to test for generalizability of their findings by studying embryonic mitosis in other organisms, and at a later stage, they want to explore the evolution of spindle assembly and embryonic divisions, which would also contribute to a better understanding of human embryogenesis and to developing diagnosis and treatment of human infertility.
“With this paper, we have created a solid foundation,” summarizes Professor Kiyomitsu, “but we have also opened a new frontier. Embryonic mitosis is beautiful, mysterious, and challenging to study, and we hope that with our work, we can eventually get a little closer to understanding the intricate processes at the beginning of life.”

Three of the researchers involved in the study, in front of fish-tanks of the transgenic medaka fish used in the study. From left to right: Dr. Ai Kiyomitsu from OIST, Professor Satoshi Ansai from Kyoto University (previously Tohoku University), and Professor Tomomi Kiyomitsu from OIST. Photo: Tomomi Kiyomitsu.
Identifying a new functions of UHRF1 to maintain DNA methylation
KanemakiGroup / Molecular Cell Engineering Laboratory
Non-canonical functions of UHRF1 maintain DNA methylation homeostasis in cancer cells
Kosuke Yamaguchi, Xiaoying Chen, Brianna Rodgers, Fumihito Miura, Pavel Bashtrykov, Frédéric Bonhomme, Catalina Salinas-Luypaert, Deis Haxholli, Nicole Gutekunst, Bihter Özdemir Aygenli, Laure Ferry, Olivier Kirsh, Marthe Laisné, Andrea Scelfo, Enes Ugur, Paola B. Arimondo, Heinrich Leonhardt, Masato T. Kanemaki, Till Bartke, Daniele Fachinetti, Albert Jeltsch, Takashi Ito & Pierre-Antoine Defossez
Nature Communications (2024) 15, 2960 DOI:10.1038/s41467-024-47314-4
Replication of the genetic material DNA is essential for cell proliferation. During this process, not only DNA but also DNA methylation, which is essential epigenetic mark, is maintained from parent to daughter cells. It has been known that DNA methyltransferase 1 (DNMT1) and its activator, UHRF1, are important to maintain DNA methylation. Although it has been recognized primarily as an activator of DNMT1, UHRF1 is also implicated in cancer development, unlike DNMT1, suggesting that it may have functions through other pathways, independent of DNMT1.
To delineate the individual functions of UHRF1 and DNMT1, we utilized the auxin-inducible degron (AID)(In Japanese only) and AID2 system that can induce the total and synchronous depletion of endogenous UHRF1 or DNMT1 proteins in human cancer cells. Interestingly, UHRF1 depletion resulted in a more severe DNA methylation loss than DNMT1 removal. With the combination of whole genome DNA methylation analysis and genetic knock-out techniques, we revealed that UHRF1 regulated not only DNMT1 but also DNA methyltransferases DNMT3A/DNMT3B and demethylase TET2.
Assistant Prof. Kosuke Yamaguchi (formally a postdoctoral researcher at Paris-Cite University) led the study as the first and co-corresponding author in the Pierre-Antoine Defossez laboratory, and Prof. Masato Kanemaki was also involved in this study.
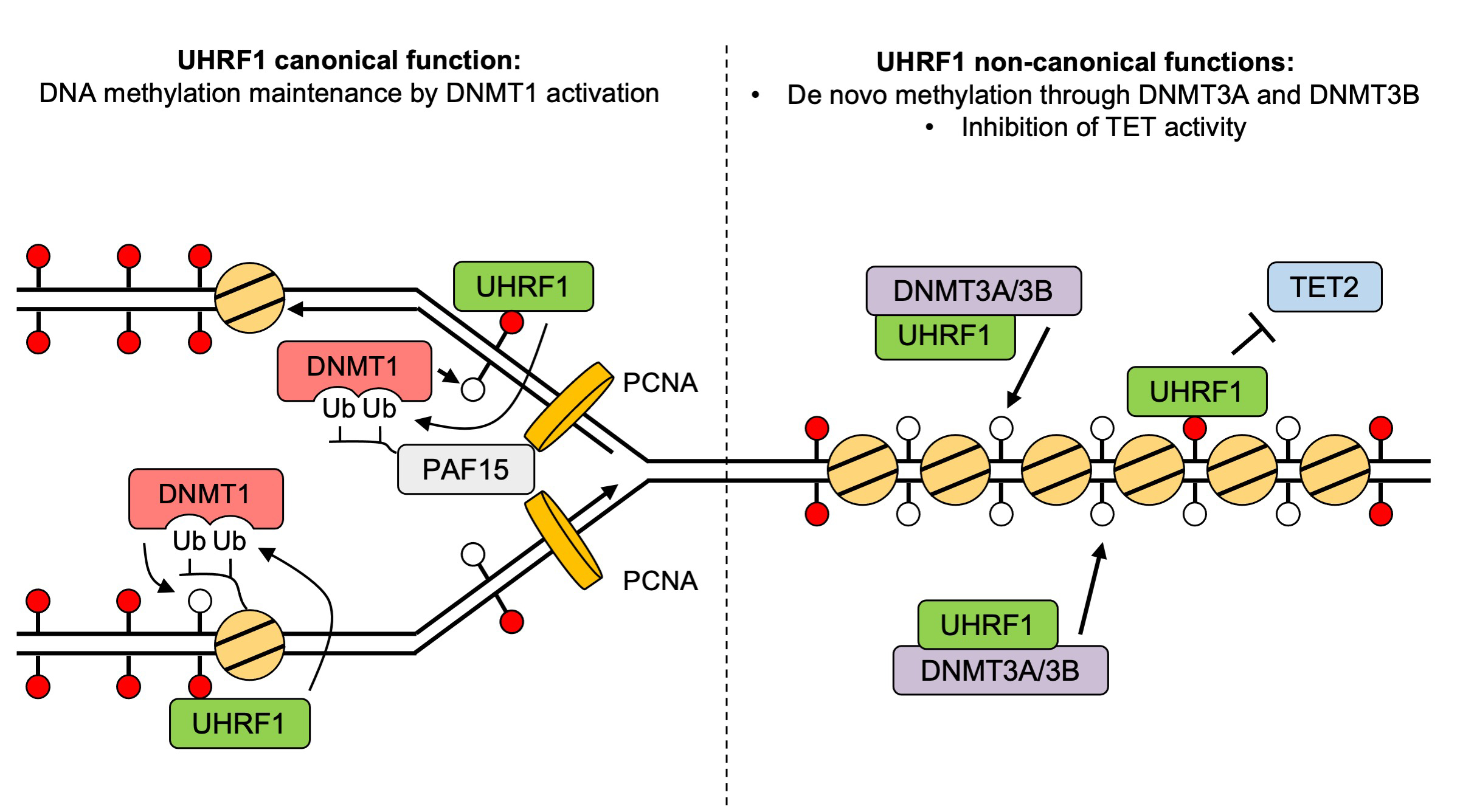
Figure: A revised and expanded model for UHRF1 functions in DNA methylation homeostasis.
Establishment of a zebrafish inbred strain, M-AB, capable of regular breeding and genetic manipulation
-Novel zebrafish bioresource from NIG-
Press release

Establishment of a zebrafish inbred strain, M-AB, capable of regular breeding and genetic manipulation
Kenichiro Sadamitsu, Fabien Velilla, Minori Shinya, Makoto Kashima, Yukiko Imai, Toshihiro Kawasaki, Kenta Watai, Miho Hosaka, Hiromi Hirata and Noriyoshi Sakai.
Scientific Reports (2024) 14, 7455 DOI:10.1038/s41598-024-57699-
![]() Press release (In Japanese only)
Press release (In Japanese only)
Inbred strains of organisms are genetically highly uniform and thus useful for life science research. We have previously reported the ongoing generation of the zebrafish IM strain from the India (IND) strain through full sib-pair mating for 16 generations. However, the IM fish laid a small number of offspring and had a short lifespan, implying the need for discreet care in breeding. Here, we report the subsequent establishment of IM strain as well as the generation of a new inbred zebrafish strain, Mishima-AB (M-AB). M-AB was derived from the *AB strain by full sib-pair mating for over 20 generations, which fulfills the general criterion for the establishment of an inbred strain. In contrast to the IM case, maintenance of the M-AB strain by sib-pair mating required almost no special handling. Genome sequencing of IM individuals from the 47th generation and M-AB individuals from the 27th generation revealed that SNP-based genomic heterogeneity across whole-genome nucleotides was 0.008% and 0.011%, respectively. These percentages were much lower than those of the parental IND (0.197%) and *AB (0.086%) strains. These results indicate that the genomes of these inbred strains were highly homogenous. We also demonstrated the successful microinjection of antisense morpholinos, CRISPR/Cas9, and foreign genes into M-AB embryos at the 1-cell stage. Overall, we report the establishment of a zebrafish inbred strain, M-AB, which is capable of regular breeding and genetic manipulation. This strain will be useful for the analysis of genetically susceptible phenotypes such as behaviors, microbiome features and drug susceptibility.
Source: Kenichiro Sadamitsu et al., Scientific Reports (2024) 14, 7455

Figure: Crossbreeding of inbred M-AB strain
A new laboratory established in the Center for Frontier Research
Dr. Kenji FUKUSHIMA joined the Center for Frontier Science as of April 1, 2024.
▶ FUKUSHIMA, Kenji : Center for Frontier Research Plant Evolution Laboratory

Associate Professor
Center for Frontier Research is an incubation center to simultaneously develop two elements: human resources and new research fields. Promising young scientists conduct research as principal investigator (tenure-track associate professor) to explore new frontiers in genetics and related areas, taking advantage of NIG’s research infrastructure and various support systems.
Three new faculty have joined NIG as of April 1, 2024
Associate Professor
Dr. Kazuhide ASAKAWA joined NIG as a Associate Professor and opened a new laboratory on April 1, 2024.
▶ ASAKAWA, Kazuhide : Neurobiology and Pathology Laboratory

ASAKAWA, Kazuhide
Associate Professor
Dr. Yasuhiro GOTO joined the Advanced Genomics Center of NIG as a Associate Professor on April 1, 2024.
▶ GOTO, Yasuhiro : Advanced Genomics Center Sequencing Division

GOTO, Yasuhiro
Associate Professor
Assistant Professor
New Assistant Professor joins NIG as of April 1, 2024.
▶ YAMAGUCHI, Kosuke : Kanemaki Group, Molecular Cell Engineering Laboratory















