Ancient Feces Reveals Intestinal Environment of Hunters-Gatherers, Jomon People
~Metagenomic analysis of human coprolites~
Press release
Metagenomic analyses of 7000 to 5500 years old coprolites excavated from the Torihama shell-mound site in the Japanese archipelago
Luca Nishimura, Akio Tanino, Mayumi Ajimoto, Takafumi Katsumura, Motoyuki Ogawa, Kae Koganebuchi, Daisuke Waku, Masahiko Kumagai, Ryota Sugimoto, Hirofumi Nakaoka, Hiroki Oota, Ituro Inoue
PLOS ONE (2024) 19, e0295924 DOI:10.1371/journal.pone.0295924
![]() Press release (In Japanese only)
Press release (In Japanese only)
The characteristics of the intestinal environment of the Jomon people who lived in the Japanese archipelago thousands of years ago were not known.
In this study, we conducted the first metagenomic analysis in Japan using ancient DNA extracted from four Jomon-period feces fossil (coprolite) samples. Analysis of the large scale DNA data obtained by metagenomic analysis revealed genome sequences derived from bacteria and viruses that are presumed to have existed in the intestines. The results of this analysis may reflect the characteristics of the Jomon people’s intestinal environment.
We plan to conduct similar analyses on other Jomon coprolites to identify more bacteria and viruses, and to elucidate the evolution of intestinal bacteria and viruses from the Jomon period to the present day, as well as the detailed characteristics of the Jomon intestinal environment.
The research was conducted by a joint research group including Luca Nishimura (a graduate student at SOKENDAI) and Project Professor Ituro Inoue from the National Institute of Genetics, Professor Hiroki Oota from Graduate School of Science, The University of Tokyo, and Mayumi Ajimoto, a curator at the Wakasa History Museum in Fukui Prefecture.
The research results were published in the international scientific journal PLOS ONE on January 25, 2024 (JST).
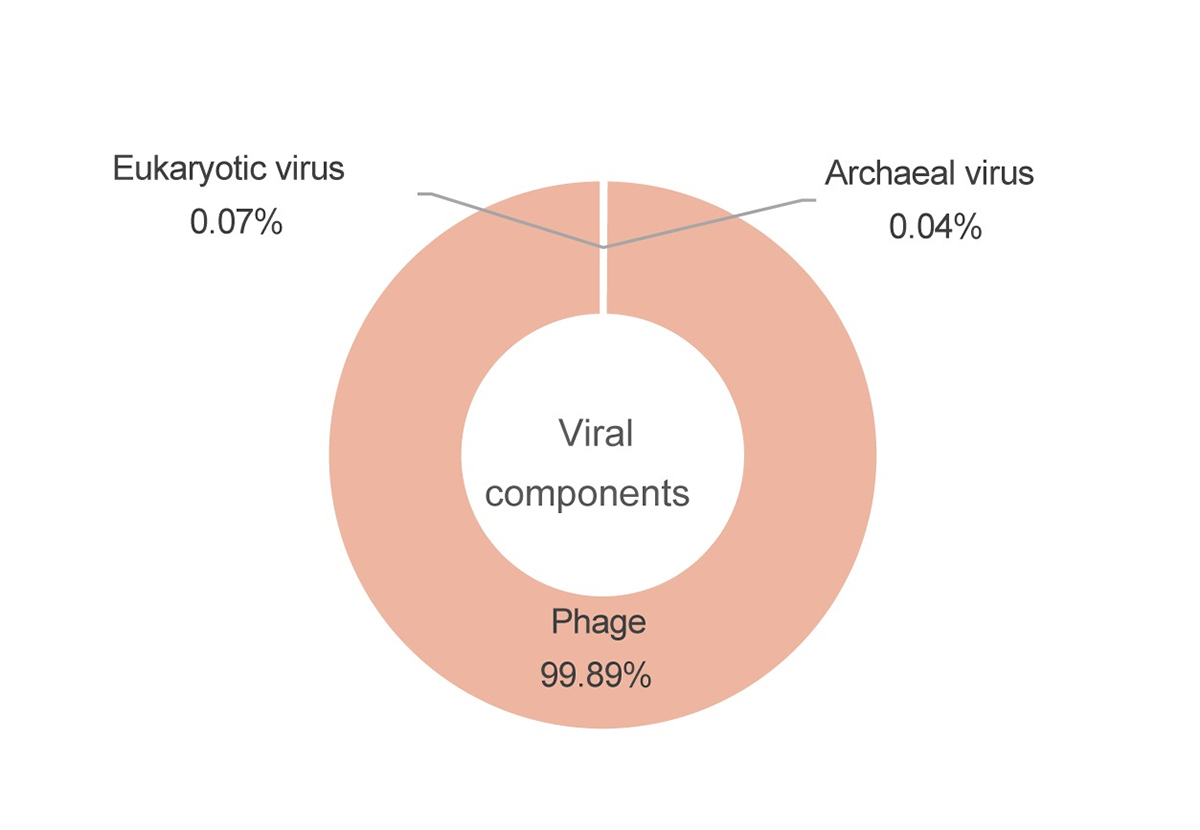
Figure: Classification of viral genome sequences detected in the Jomon coprolites.
The pie chart shows the ratio of virus from eukaryotes as 0.07%, archaea as 0.04%, and viruses infecting bacteria (phage), as 99.89%. You can see that most of the viruses are phages that















