A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Nakamura Group / Genome Informatics Laboratory
A Practical Assembly Guideline for Genomes with Various Levels of Heterozygosity
Takako Mochizuki, Mika Sakamoto, Yasuhiro Tanizawa, Takuro Nakayama, Goro Tanifuji, Ryoma Kamikawa, Yasukazu Nakamura*
*Corresponding Author
Briefings in Bioinformatics (2023) 24, bbad337 DOI:10.1093/bib/bbad337
The advancement of long-read sequencing technologies, exemplified by subreads of Pacific Biosciences, has significantly advanced our ability to reconstruct genome sequences. While these technologies can generate long reads, they are plagued by high sequence errors. To address these errors and strive to construct long, highly accurate contig sets, various de novo assemblers have been developed.
In de novo assembly of diploid genomes, the complexity increases with higher heterozygosity. Therefore, heterozygosity is a significant factor influencing the completeness of de novo assembly. However, systematic evaluations of de novo assemblers for diploid genomes with various heterozygosity levels have not been conducted.
In this study, using genomes with varying levels of heterozygosity, we conducted a series of processes, including estimation of genome characteristics such as genome size and heterozygosity, de novo assembly, polishing, and removing contigs including alleles. We have presented a guideline for constructing a representative haplotype set based on heterozygosity levels.
This work was supported by JSPS grant-in-aid for Scientific Research on Innovative Areas, Platform for Advanced Genome Science [16H06279], and KAKENHI [15H05606 and 19H03274, 20H03305, 17H03723].
Computations were partially performed on the National Institute of Genetics supercomputer.
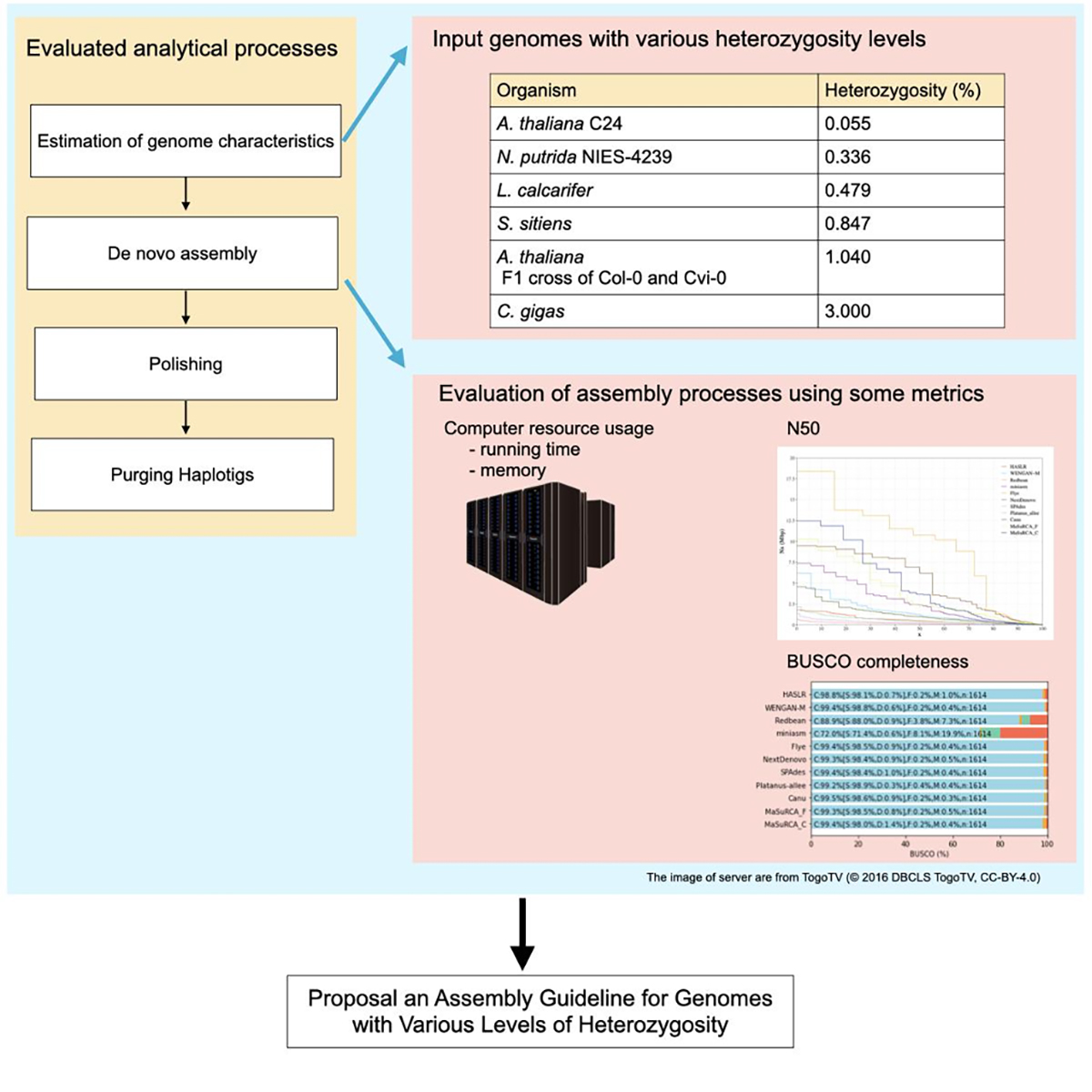
Figure: An evaluation process for genome assembly using genomes with various levels of heterozygosity















