POLR1A variant- and tissue-specific effects underlie phenotypic heterogeneity in craniofacial, neural and cardiac anomalies
POLR1A variant- and tissue-specific effects underlie phenotypic heterogeneity in craniofacial, neural and cardiac anomalies
Kelly Smallwood, Kristin E.N. Watt, Satoru Ide, Kristina Baltrunaite, Chad Brunswick, Katherine Inskeep, Corrine Capannari, Margaret P. Ada, Amber Begtrup, Debora R. Bertola, Laurie Demmer, Erin Demo, Orrin Devinsky, Emily R. Gallagher, Maria J. Guillen Sacoto, Robert Jech, Boris Keren, Jennifer Kussmann, Roger Ladda, Lisa A. Lansdon, Sebastian Lunke, Anne Mardy, Kirsty McWalters, Richard Person, Laura Raiti, Noriko Saitoh, Carol J. Saunders, Rhonda Schnur, Matej Skorvanek, Susan L. Sell, Anne Slavotinek, Bonnie R. Sullivan, Zornitza Stark, Joseph D. Symonds, Tara Wenger, Sacha Weber, Sandra Whalen, Susan M. White, Juliane Winkelmann, Michael Zech, Shimriet Zeidler, Kazuhiro Maeshima, Rolf W. Stottmann, Paul A Trainor, K. Nicole Weaver
The American Journal of Human Genetics (2023) 110, 809-825 DOI:10.1016/j.ajhg.2023.03.014
Heterozygous pathogenic variants in POLR1A, which encodes the largest subunit of RNA Polymerase I, were previously identified as the cause of Acrofacial Dysostosis, Cincinnati-type. The predominant phenotypes observed in the initial cohort of 3 individuals were craniofacial anomalies reminiscent of Treacher Collins syndrome. We subsequently identified 17 additional individuals with 12 unique (11 novel) heterozygous variants in POLR1A and observed numerous additional phenotypes including neurodevelopmental abnormalities and structural cardiac defects, in combination with highly prevalent craniofacial anomalies and variable limb defects. To understand the pathogenesis of this pleiotropy, we modeled an allelic series of POLR1A variants in vitro and in vivo. In vitro assessments demonstrate variable effects of individual pathogenic variants on ribosomal RNA synthesis and nucleolar morphology which supports the possibility of variant-specific phenotypic effects in patients. To further explore variant-specific effects in vivo, we used CRISPR/Cas9 gene editing to recapitulate two human variants in mice. Additionally, spatiotemporal requirements for Polr1a in developmental lineages contributing to congenital anomalies in patients were examined via conditional mutagenesis in neural crest cells (face and heart), the second heart field (cardiac outflow tract and right ventricle), and forebrain precursors in mice (Figure A). Consistent with its ubiquitous role in the essential function of ribosome biogenesis, we observed that loss of Polr1a in any of these lineages causes death of the affected cells and resulting embryonic malformations. Altogether, our work greatly expands the phenotype of human POLR1A-related disorders and demonstrates variant-specific effects and differential tissue-specific requirements that provide novel insights into the underlying pathogenesis of ribosomopathies.
Assistant Professor Satoru Ide and Professor Kazuhiro Maeshima of Genome Dynamics Laboratory performed in vitro assessment of POLR1A variants, which impact rRNA transcription and the nucleolar morphology in human culture cells expressing wild-type or mutant POLR1A. Using a machine learning algorithm called “wndchrm”, Ide and Maeshima found that nucleolar morphologies of the mutants (based on POLR1A foci) are somehow corelated with their rRNA transcription capacities or status (Figure B).
The part to which NIG contributed was supported by the Japan Society for the Promotion of Science (JSPS) grants (22H05606, 21H02535 to SI; 19H05273 ,20H05936 to KM) etc. The image analysis by the machine learning algorithm “wndchrm” was conducted in collaboration with Dr. Noriko Saito in The Cancer Institute of JFCR.
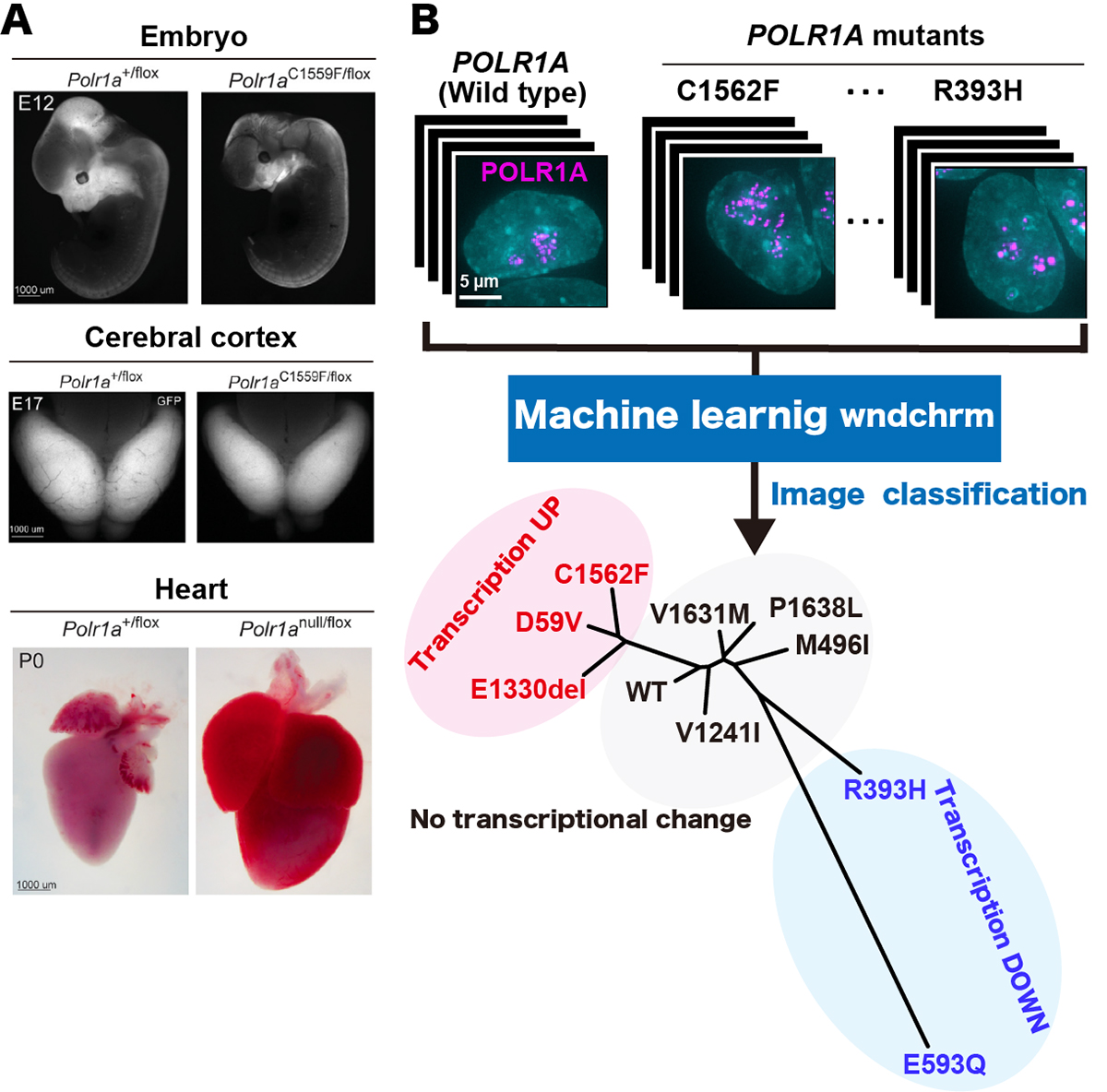
Figure: (A)Polr1a variant murine embryos demonstrate more hypoplastic craniofacial primordia at E12 (upper panel, right), a reduced cerebral cortex area at E12 (middle panel, right) and gross enlargement of heart at P0 (lower panel, right), compared to wild type (all the panels, left), respectively. (B) (upper panel) Localization of fluorescently labeled POLR1A (cellular nucleus, cyan and human POLR1A variants, magenta). (lower panel) Phylogeny shows the similarity of the nucleolar morphologies of wild type and nine POLR1A variants. POLR1A variants showing transcription upregulation, transcription downregulation and no transcriptional change are indicated in red, blue, and black, respectively.















