Chromatin organization and DNA damage
Maeshima Group / Genome Dynamics Laboratory
Chromatin organization and DNA damage.
K. Minami, S. Iida, K. Maeshima
The Enzymes “DNA Damage and Double Strand Breaks” 2022 September 27 DOI:10.1016/bs.enz.2022.08.003
Free link (until 2022.11.16)
Genomic DNA is organized three-dimensionally in the nucleus as chromatin. Recent accumulating evidence has demonstrated that chromatin organizes into numerous dynamic domains in higher eukaryotic cells, which act as functional units of the genome. However, the cellular genome is constantly threatened by many sources of DNA damage (e.g., radiation). How do cells maintain their genome integrity when subjected to DNA damage?
In this review, we discuss how the compact state of chromatin safeguards the genome from radiation damage and chemical attacks. Together with recent genomics data, our finding (Takata et al. “Chromatin compaction protects genomic DNA from radiation damage”. PLOS ONE (2013). DOI: 10.1371/journal.pone.0075622) suggests that DNA compaction, such as chromatin domain formation, plays a critical role in maintaining genome integrity. But does the formation of such domains limit DNA accessibility inside the domain and hinder the recruitment of repair machinery to the damaged site(s) during DNA repair? To approach this issue, we first describe a sensitive imaging method to detect changes in chromatin states in living cells (single-nucleosome imaging). We then use this method to explain how cells can overcome potential recruiting difficulties; cells can decompact chromatin domains following DNA damage and temporarily increase chromatin motion (∼ DNA accessibility) to perform efficient DNA repair (Iida et al. “Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells” Science Advances (2022). DOI: 10.1126/sciadv.abn5626).
This review article will be published as the 3rd chapter in the book “The Enzymes vol.51: DNA Damage and Double Strand Breaks (Elsevier, 2022)”. This work was supported by JSPS and MEXT KAKENHI grants (19H05273 and 20H05936 to K. Maeshima), the Takeda Science Foundation (to K. Maeshima), and the Uehara Memorial Foundation (to K. Maeshima). K. Minami and S. Iida are SOKENDAI Special Researchers supported by JST SPRING JPMJSP2104.
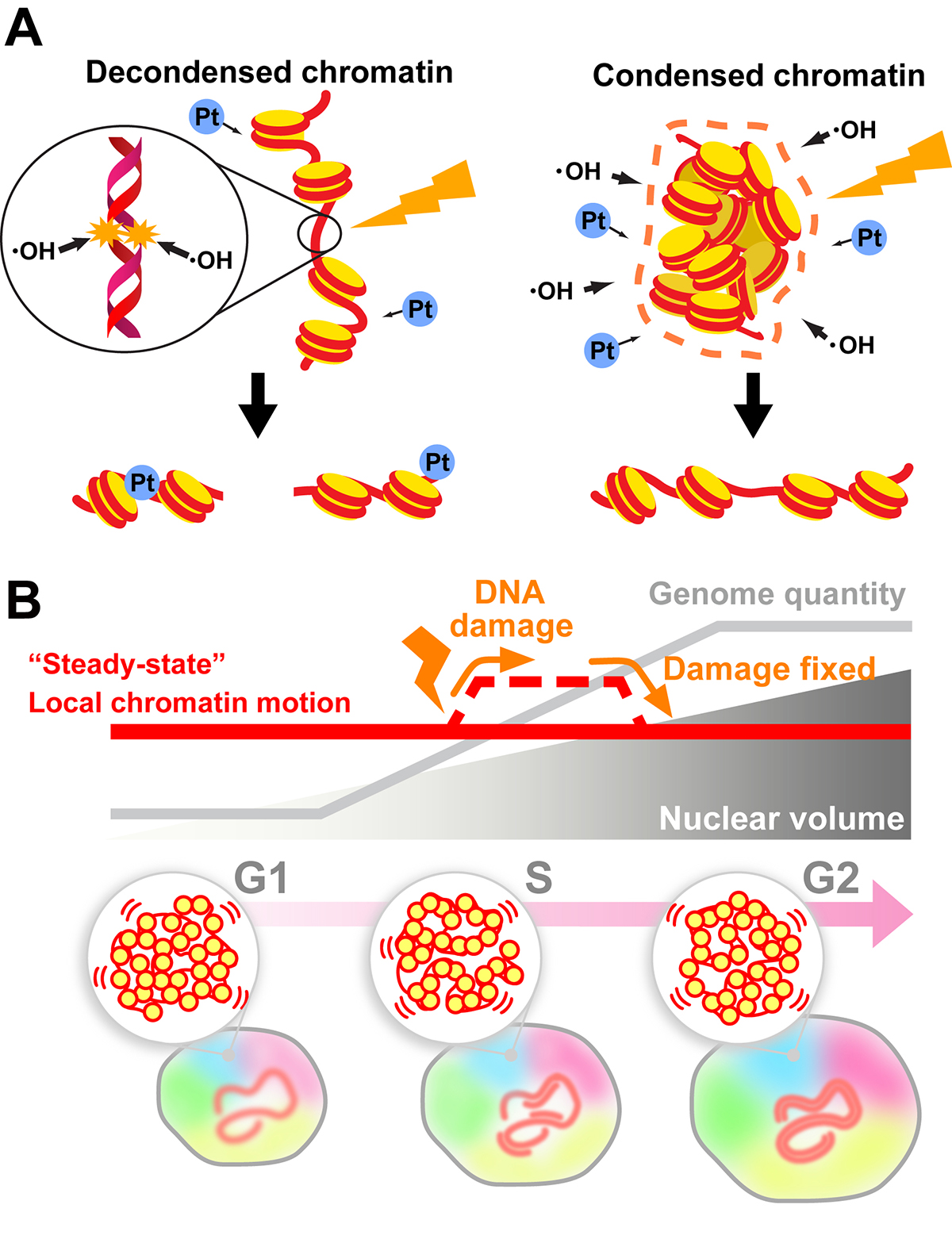
(B) Local chromatin motion persisted throughout interphase of the cell cycle (“steady-state” local chromatin motion; red line) can be transiently increased by a DNA damage response (dashed line). This temporal change in chromatin motion presumably occurs to facilitate access for DNA repair machinery.















