Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
Maeshima Group / Genome Dynamics Laboratory
Chromatin behavior in living cells: lessons from single-nucleosome imaging and tracking
*Satoru Ide, Sachiko Tamura, *Kazuhiro Maeshima *corresponding author
BioEssays 2022 June 03 DOI:10.1002/bies.202200043
Eukaryotic genome DNA is wrapped around core histones and forms a nucleosome structure. Together with associated proteins and RNAs, these nucleosomes are organized three-dimensionally in the cell as chromatin. Emerging evidence demonstrates that chromatin consists of rather irregular and variable nucleosome arrangements without the regular fiber structure and that its dynamic behavior plays a critical role in regulating various genome functions. Single-nucleosome imaging is a promising method to investigate chromatin behavior in living cells. It reveals local chromatin motion, which reflects chromatin organization not observed in chemically fixed cells. The motion data is like a gold mine. Data analyses from many aspects bring us more and more information that contributes to better understanding of genome functions. In this review article, we describe imaging of single-nucleosomes and their tracked behavior through oblique illumination microscopy. We also discuss applications of this technique, especially in elucidating nucleolar organization in living cells.
This work was supported by JSPS Grants and MEXT KAKENHI grants (21H02535, 20H05936 and 21H02453) and the Uehara Memorial Foundation.
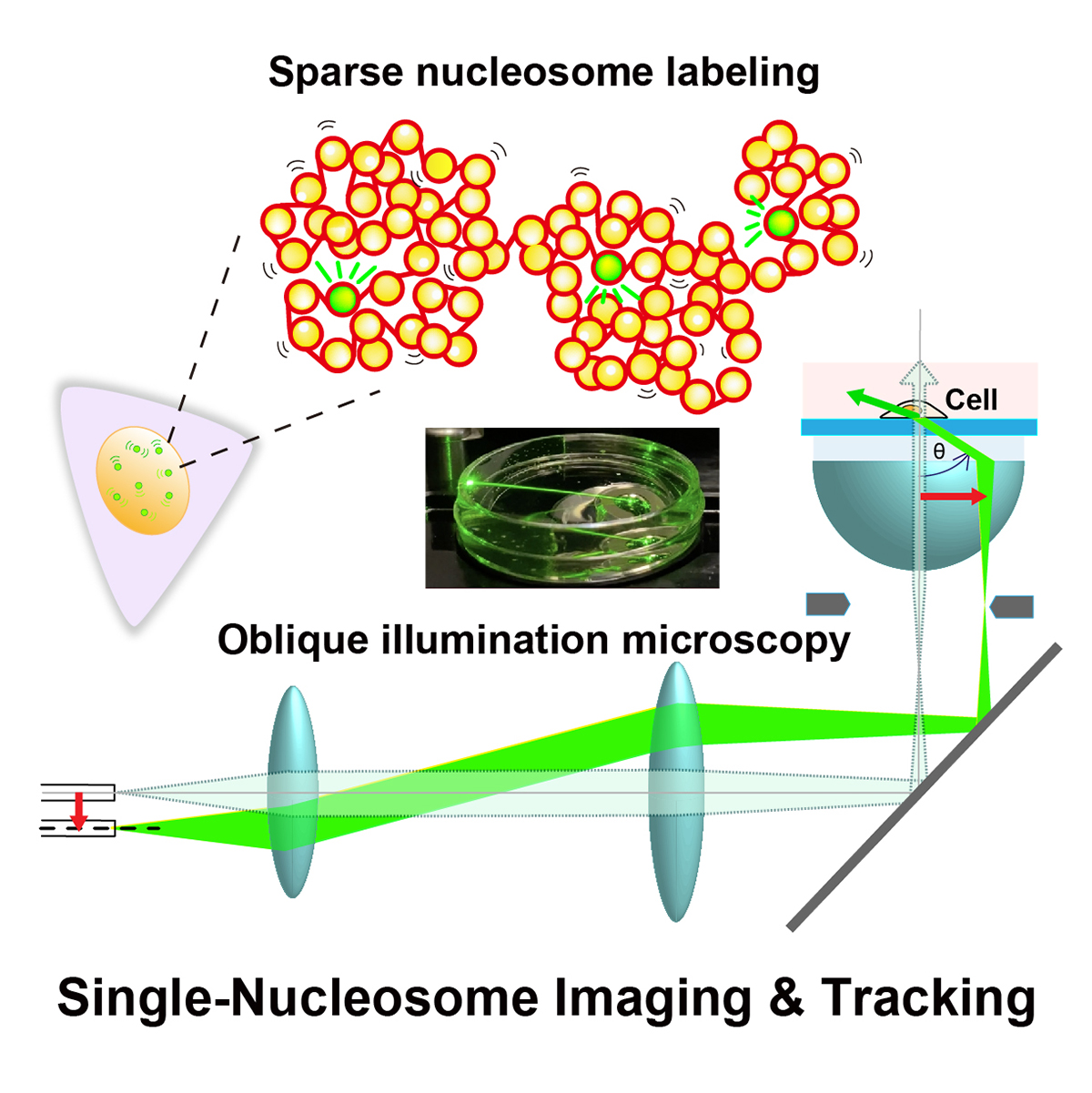
Figure: Single-nucleosome imaging and tracking is a promising technique to investigate dynamic chromatin behavior in living cells. We describe how this imaging works using sparse nucleosome labeling and oblique illumination microscopy, and how information on chromatin dynamics can be extracted from the obtained motion data.
Video1: How laser beams traveled through a glass dish filled with medium using an oblique illumination microscopy. As the incident laser beam shifts from the center axis, higher refraction angles of the beam from the glass are obtained. At the last 2 frames, total internal reflection (TIR) occurs and only the glass surface is illuminated.
Video2: Movie data (50 ms/frame) of single nucleosomes (basic units of chromatin) fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.















