How do genes move in living cells?
Press release
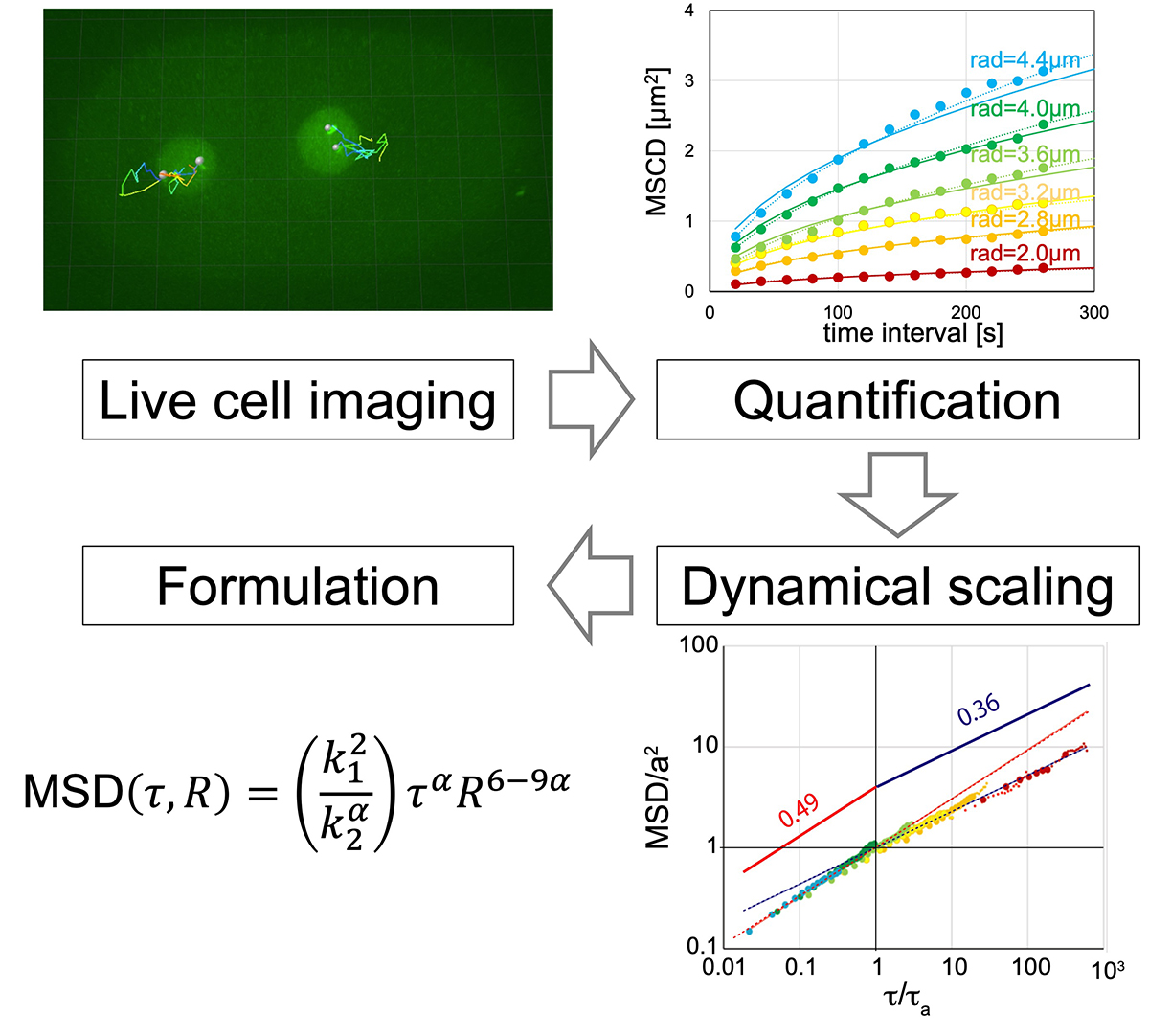
Formulation of chromatin mobility as a function of nuclear size during C. elegans embryogenesis using polymer physics theories.
Aiya K. Yesbolatova, Ritsuko Arai, *Takahiro Sakaue, and *Akatsuki Kimura. *Corresponding authors
Physical Review Letters (2022) 128, 178101 DOI:10.1103/PhysRevLett.128.178101
![]() Press release (In Japanese only)
Press release (In Japanese only)
By combining theory and experiment, Aiya K. Yesbolatova and her collaborators provided unambiguous evidence that the chromatin in early embryos obeys the universal dynamics predicted by the polymer physics. Although the chromatin dynamics is believed to play a key role in controlling the gene expression, its quantitative characterization has been elusive mainly due to the complexity in living cells. The findings and formulation help researchers quantify the chromatin motion in living cells, thus laying the foundation for future research in this biologically important problem.