Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Functional diversity of gibberellin activation enzymes leading to interspecific differences in pollen tube germination/elongation among Oryza species
Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza
Kyosuke Kawai, Sayaka Takehara, Toru Kashio, Minami Morii, Akihiko Sugihara, Hisako Yoshimura, Aya Ito, Masako Hattori, Yosuke Toda, Mikiko Kojima, Yumiko Takebayashi, Hiroyasu Furuumi, Ken-ichi Nonomura, Bunzo Mikami, Takashi Akagi, Hitoshi Sakakibara, Hidemi Kitano, Makoto Matsuoka & Miyako Ueguchi-Tanaka
Communications Biology (2022) 5, 67 DOI:10.1038/s42003-022-03008-5
The plant hormone gibberellin (GA) plays important roles in various developmental events, such as stem elongation, induction of seed germination and flowering. Although GA is indispensable for anther and pollen development, our knowledge of GA functions during plant reproduction has been limited to date.
This paper reports the functional and evolutionary analyses of rice gibberellin 3-oxidase 1 (OsGA3ox1), a gibberellin synthetic enzyme specifically expressed in the late developmental stages of anthers. Enzymatic and X-ray crystallography analyses reveal that OsGA3ox1 has a higher GA7 synthesis ratio than OsGA3ox2. In addition, we generate an osga3ox1 knockout mutant by genome editing and demonstrate the bioactive gibberellic acid synthesis by the OsGA3ox1 action during starch accumulation in pollen via invertase regulation. Furthermore, we analyze the evolution of Oryza GA3ox1s and reveal that their enzyme activity and gene expression have evolved in a way that is characteristic of the Oryza genus and contribute to their male reproduction ability.
In this paper, we used the wild strains of genus Oryza that NIG has conserved with the support of National Bioresource Project (NBRP) Rice, Ministry of Education, Culture, Sports, Science and Texhnology (MEXT), Japan.
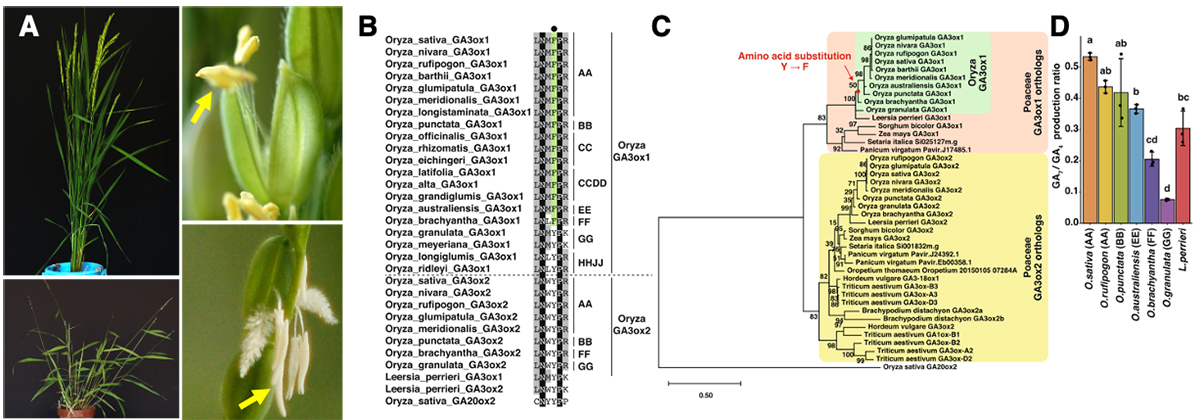
(A) Plant types and florets of cultivated species, Oryza sativa (top), and wild relatives, O. granulata (bottom). Yellow arrows indicates anthers. (B) Amino acid sequence alignment in the 2OG-interacting site around F (in OsGA3ox1) of Poaceae GA3ox orthologs and OsGA20ox2. Residues of F in the interacting site of 2OG are indicated in green, those that are 100% identical are shown in black, and those with more than 50% identity are depicted in gray. The black dot indicates the amino acid in the interacting site with co-substrate 2OG, F (shown in green), or Y. On the right side of the alignment, the letters AA and BB indicate the AA and BB Oryza genomes, respectively. (C) Phylogenetic tree of Poaceae GA3ox orthologs and OsGA20ox2. The red dot indicates the common ancestor of O. sativa to O. brachyantha that acquired the residue F in the active site. (D) In vitro enzyme activity of GA3ox1 of Oryza species and L. perrieri; O. sativa GA3ox1, O. rufipogon GA3ox1, O. punctata GA3ox1, O. brachyantha GA3ox1, O. granulata GA3ox1, L. perrieri GA3ox1. Error bars, s.d. n = 3, one-way ANOVA with Tukey’s multiple comparisons test. Different letters denote significant differences (p < 0.05).
「Quantitative Biology – A practical introduction」from Springer by Professor Akatsuki KIMURA

Professor Akatsuki Kimura has published an English textbook “Quantitative Biology – A practical introduction” from Springer.
The textbook is based on his lecture on quantitative biology at SOKENDAI, including the codes of computer programming.
Target reader is a complete beginner of computer programming.
The textbook also overviews “Cell Architectonics”, a cell biology research Prof. Kimura is pursuing.
- title: Quantitative Biology – A practical introduction
Mr. Harsha Somashekar received the Best Papers Award 2021 in the 93rd Annual Meeting of the Genetics Society of Japan.
Mr. Harsha Somashekar (Plant Cytogenetics/Nonomura Lab), a PhD student (D4) in the graduate university SOKENDAI, received the Best Papers Award 2021 in the 93rd Annual Meeting of the Genetics Society of Japan.

Title:
Hyper accumulation of callose at extracellular spaces of anther locules is required for normal progression of male meiosis in rice.
A new method of rapid protein depletion in nematode individuals using an improved auxin-inducible degron system, AID2
The auxin-inducible degron 2 (AID2) system enables controlled protein knockdown during embryogenesis and development in Caenorhabditis elegans.
Negishi T#, Kitagawa S#, Horii N, Tanaka Y, Haruta N, Sugimoto A, Sawa H, Hayashi KI, Harata M*, Kanemaki MT*.
# These authors contributed equally * Co-corresponding authors
Genetics (2022) 220, iyab218 DOI:10.1093/genetics/iyab218
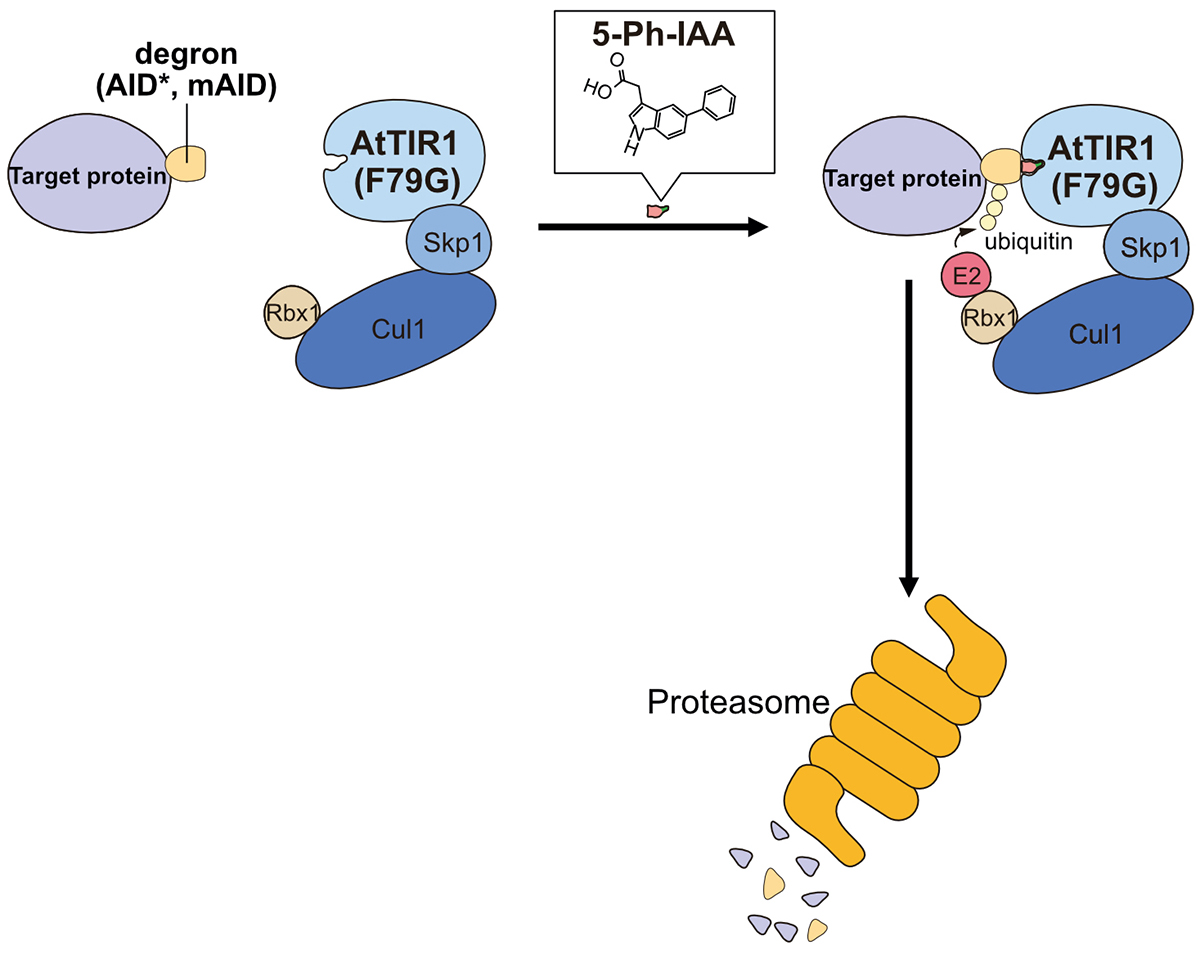
To analyze the protein function of C. elegans, it is useful to study the phenotype by suppressing the protein function. For this purpose, nematode individuals with mutated genes and the RNA interference method have been used. However, genes that play an essential role in nematode development may cause a developmental defect due to the loss of the gene, making further analysis difficult. In addition, due to a large amount of maternally-derived mRNA and proteins in the eggs, the functions of proteins normally involved in early development may not be expressed as a phenotype in early development using existing techniques. The auxin-inducible degron (AID) system that we have established rapidly degrades and removes target proteins at any given time and allows us to observe phenotypes that cannot be seen using existing techniques. The AID method has already been applied to C. elegans by a group in the U.S. and is now being used in various studies. However, the conventional AID method has some problems, such as leaky target degradation in the absence of auxin and the effects of high concentrations of auxin.
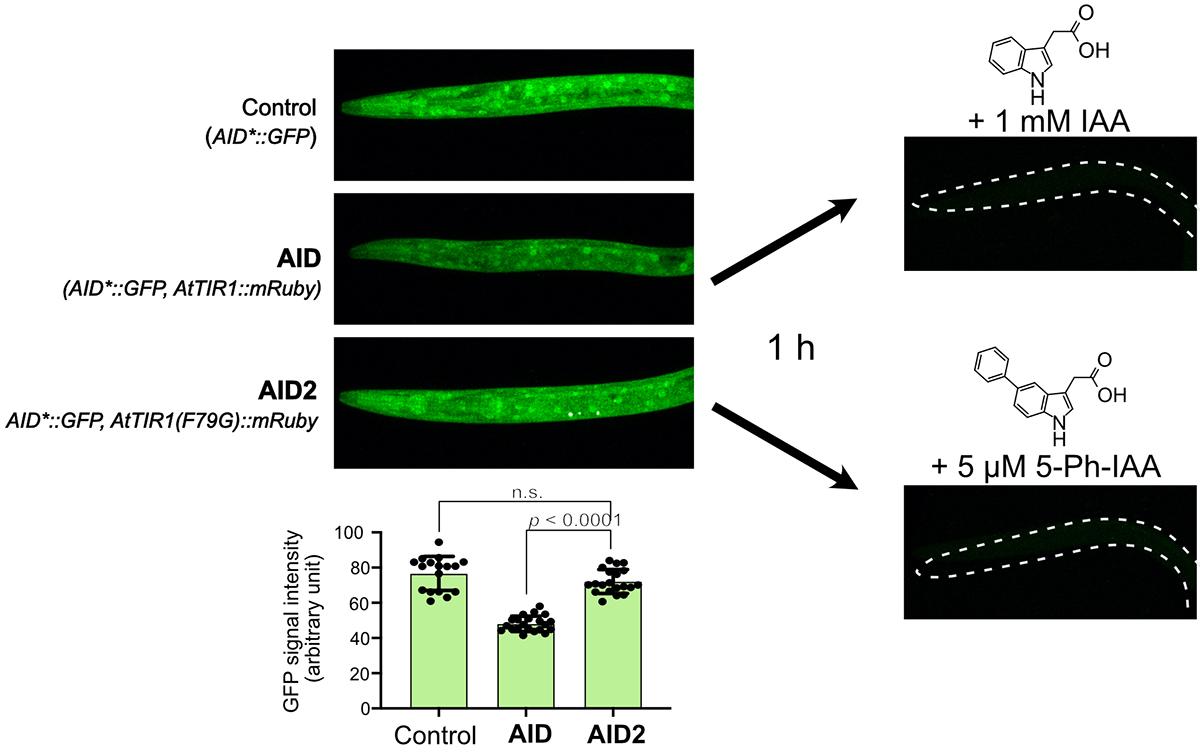
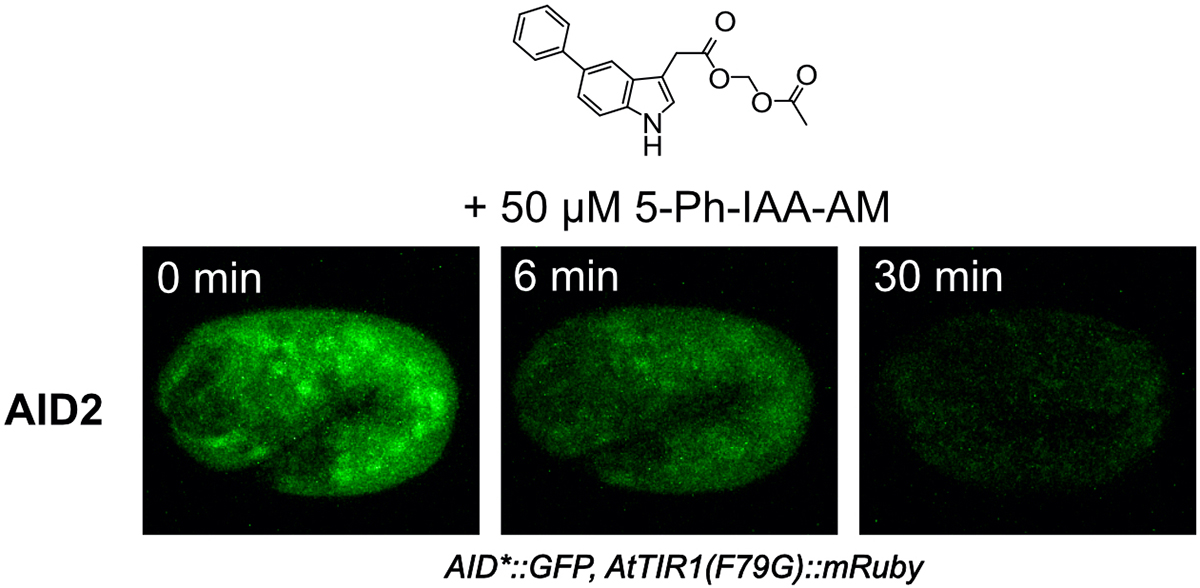
Therefore, we applied an improved method, AID2, which we developed last year using budding yeast, cultured cells, and mice, to C. elegans to overcome these problems (Figure 1). As a result, we found that AID2 completely suppressed the leaky degradation in C. elegans and rapidly induced target degradation with 1/1300 of the ligand concentration (Figure 2). Furthermore, by degrading the histone H2A.Z protein, we succeeded in observing a developmental defect that had not been reported previously. Furthermore, to induce degradation in the embryo in the egg, we developed a modified ligand suitable for eggshell permeabilization, enabling rapid proteolysis in the embryo (Figure 3).
This research was led by Assistant Professor Takefumi Negishi and Professor Masato Kanemaki at the National Institute of Genetics, and a graduate student Saho Kitagawa and Professor Masahiko Harada at Tohoku University, in collaboration with Professor Kenichiro Hayashi at Okayama University of Science, Professor Hitoshi Sawa at the National Institute of Genetics, and Professor Asako Sugimoto at Tohoku University.