Can genomics disentangle the enchanting mechanisms of shark and ray morphogenesis?
Kuraku Group / Molecular Life History Laboratory
Shark and ray genomics for disentangling their morphological diversity and vertebrate evolution
Shigehiro Kuraku
Developmental Biology 477 262-272 (2021) DOI:10.1016/j.ydbio.2021.06.001
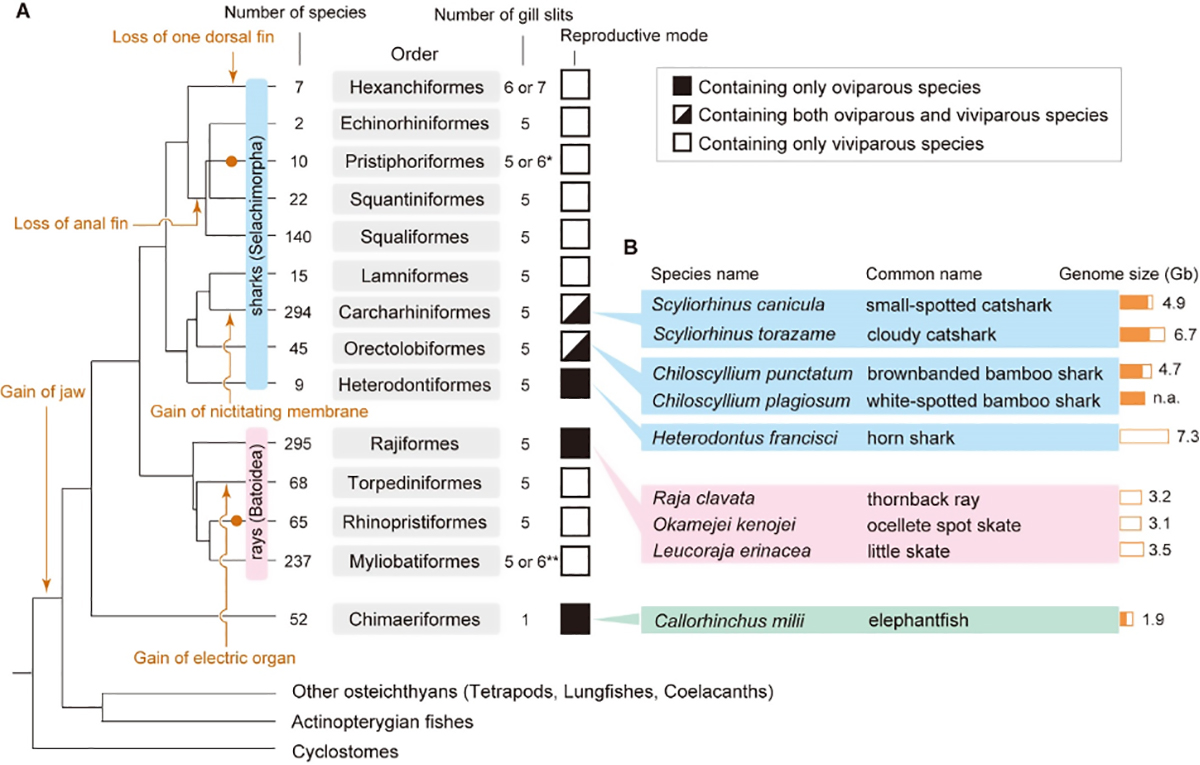
Developmental studies of sharks and rays (elasmobranchs) have provided much insight into the process of morphological evolution of vertebrates. Although those studies are supposedly fueled by large-scale molecular sequencing information, whole-genome sequences of sharks and rays were made available only recently. One compelling difficulty of elasmobranch developmental biology is the low accessibility to embryonic study materials and their slow development. Another limiting factor is the relatively large size of their genomes. Moreover, their large body sizes restrict sustainable captive breeding, while their high body fluid osmolarity prevents reproducible cell culturing for in vitro experimentation, which has also limited our knowledge of their chromosomal organization for validation of genome sequencing products. This article focuses on egg-laying elasmobranch species used in developmental biology and provides an overview of the characteristics of the shark and ray genomes revealed to date.