Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Inner-dual structure of modern Yaponesians (people in Japanese Archipelago)
Press release
Genome-wide SNP data of Izumo and Makurazaki populations support inner-dual structure model for origin of Yamato people
Timothy Jinam, Yosuke Kawai, Yoichiro Kamatani, Shunro Sonoda, Kanro Makisumi, Hideya Sameshima, Katsushi Tokunaga, and Naruya Saitou
Journal of Human Genetics 2021 January 25. DOI:10.1038/s10038-020-00898-3
Press release (In Japanese only)
The “Dual Structure” model on the formation of the modern Japanese population assumes that the indigenous hunter- gathering population (symbolized as Jomon people) admixed with rice-farming population (symbolized as Yayoi people) who migrated from the Asian continent after the Yayoi period started. The Jomon component remained high both in Ainu and Okinawa people who mainly reside in northern and southern Japan, respectively, while the Yayoi component is higher in the mainland Japanese (Yamato people). The model has been well supported by genetic data, but the Yamato population was mostly represented by people from Tokyo area. We generated new genome-wide SNP data using Japonica Array for 45 individuals in Izumo City of Shimane Prefecture and for 72 individuals in Makurazaki City of Kagoshima Prefecture in Southern Kyushu, and compared these data with those of other human populations in East Asia, including BioBank Japan data. Using principal component analysis, phylogenetic network, and f4 tests, we found that Izumo, Makurazaki, and Tohoku populations are slightly differentiated from Kanto (including Tokyo), Tokai, and Kinki regions. These results suggest the substructure within Mainland Japanese maybe caused by multiple migration events from the Asian continent following the Jomon period, and we propose a modified version of “Dual Structure” model called the “Inner-Dual Structure” model.
This study was supported by SOKENDAI Cooperative Research grant on modern human evolution, MEXT Grant-in-aid for Scientific Research on Innovative Areas “Yaponesian Genome” (grant number 18H05505), cooperative research of the Inter-University Research Institutions (grant number I- URIC18P01), and a cooperative research with Genesis Healthcare.
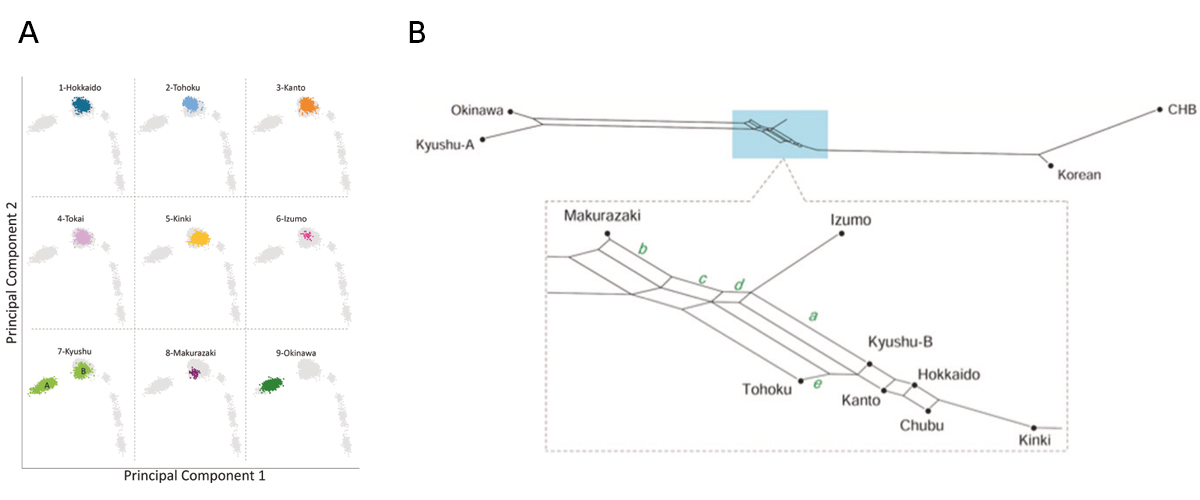
Figure: Genetic relationship of nine Yaponesian populations and some continental populations.
(A) Principal Component Analysis (PCA). DNA data for Izumo (6) and Makurazaki (8) are newly determined, and those for other seven regions are from BioBank Japan. Five continental populations are located at lower right side. Kyushu (7) population is divided into Okinawa cluster A and Yamato cluster B. (B) Phylogenetic network analysis using the Neighbor-Net method. Okinawa and Kyushu-A are located at left and Korean and CHB (Chinese Han in Beijing) are located at right. Makurazaki is the closest to the left cluster followed by Izumo, while Kinki is the closest to the right cluster.
Non-thalamic origin of zebrafish sensory nuclei implies convergent evolution of visual pathways in amniotes and teleosts
Non-thalamic origin of zebrafish sensory nuclei implies convergent evolution of visual pathways in amniotes and teleosts
Solal Bloch, Hanako Hagio, Manon Thomas, Aure´ lie Heuze´, Jean-Michel Hermel, Elodie Lasserre, Ingrid Colin, Kimiko Saka, Pierre Affaticati, Arnim Jenett, Koichi Kawakami, Naoyuki Yamamoto, Kei Yamamoto
Elife 9, e54945 (2020) DOI:10.7554/eLife.54945
Ascending visual projections similar to the mammalian thalamocortical pathway are found in a wide range of vertebrate species, but their homology is debated. To get better insights into their evolutionary origin, we examined the developmental origin of a thalamic-like sensory structure of teleosts, the preglomerular complex (PG), focusing on the visual projection neurons. Similarly to the tectofugal thalamic nuclei in amniotes, the lateral nucleus of PG receives tectal information and projects to the pallium. However, our cell lineage study in zebrafish reveals that the majority of PG cells are derived from the midbrain, unlike the amniote thalamus. We also demonstrate that the PG projection neurons develop gradually until late juvenile stages. Our data suggest that teleost PG, as a whole, is not homologous to the amniote thalamus. Thus, the thalamocortical-like projections evolved from a non-forebrain cell population, which indicates a surprising degree of variation in the vertebrate sensory systems.
This study was conducted as collaboration with Dr. Kei Yamamoto at Universite´ Paris-Saclay and CNRS and Dr. Naoyuki Yamamoto at Graduate School of Bioagricultural Sciences, Nagoya University. This study was partially supported by NBRP and NBRP/Genome Information upgrading program from AMED and NIG-JOINT (2013-A15).
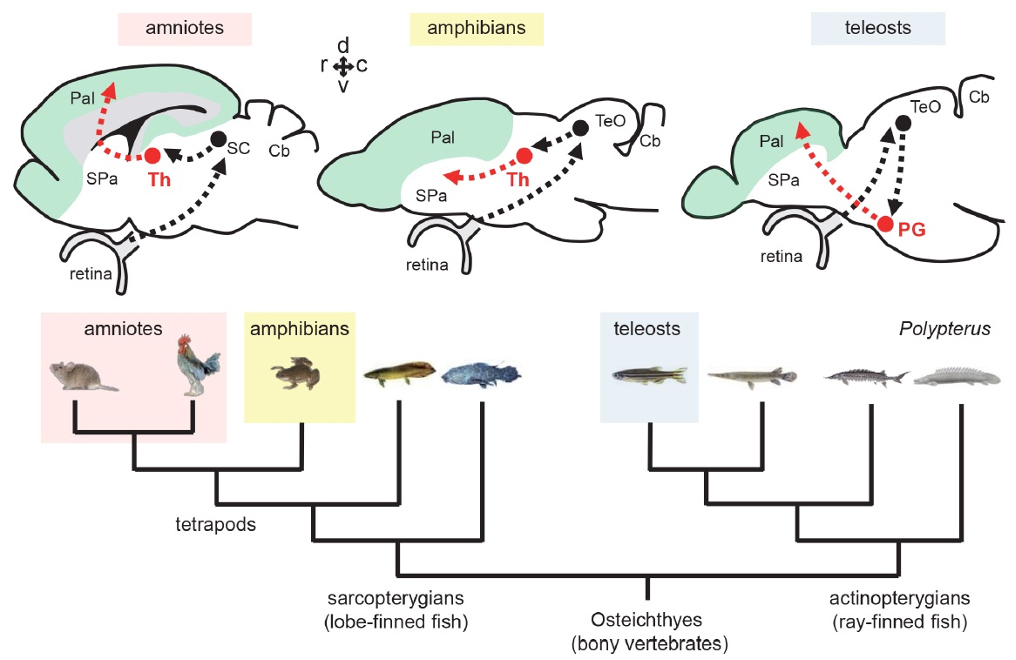
Figure1: Tectofugal pathways in amniotes, amphibians, and teleosts and their phylogenetic relationships.
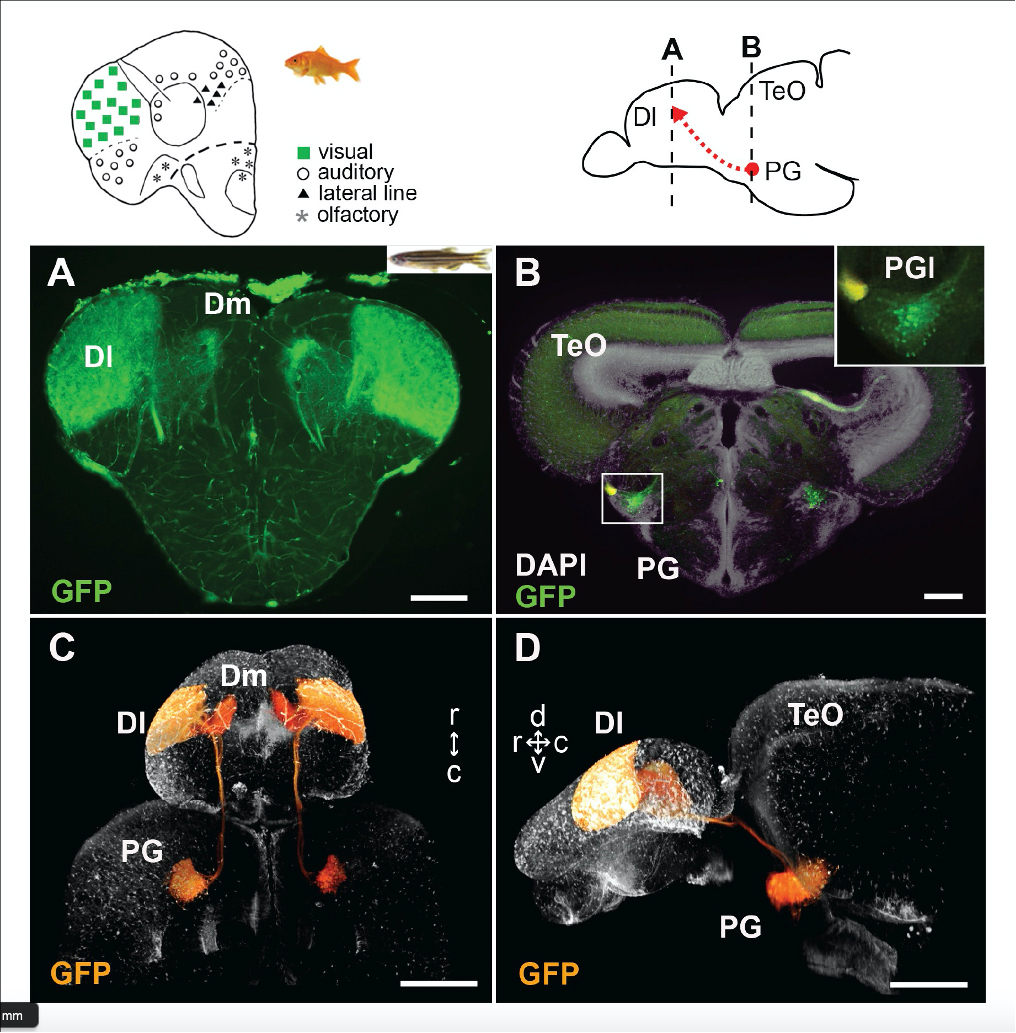
Figure2: Visualization of the preglomerular complex (PG) cells and their projection to the pallium in gene-trap transgenic fish.
Schedule of Admissions 2021
Hexanediol immobilizes chromatin
1,6-hexanediol rapidly immobilizes and condenses chromatin in living human cells
Yuji Itoh, Shiori Iida, Sachiko Tamura, Ryosuke Nagashima, Kentaro Shiraki, Tatsuhiko Goto, Kayo Hibino, Satoru Ide and Kazuhiro Maeshima
Life Science Alliance 4, e202001005 (2021) DOI:10.26508/lsa.202001005
Liquid droplets formed inside the cell by liquid-liquid phase separation (LLPS) maintain membrane-less condensates/bodies (or compartments). These droplets are important for concentrating certain molecules and facilitating spatiotemporal regulation of cellular functions. 1,6-hexanediol (1,6-HD), an aliphatic alcohol, inhibits weak hydrophobic protein-protein/protein-RNA interactions required for the droplet formation (droplet melting activity) and is used here to elucidate the formation process of cytoplasmic/nuclear condensates/bodies. However, the effect of 1,6-HD on chromatin in living cells remains unclear. We found that 1,6-HD drastically suppresses chromatin motion and hyper-condenses chromatin in human cells by using live cell single-nucleosome imaging, which detects changes in the state of chromatin. These effects were enhanced in a dose-dependent manner. Chromatin was ‘frozen’ by 5%, or higher, concentrations of 1,6-HD. 1,6-HD greatly facilitated cation-dependent chromatin condensation in vitro. This 1,6-HD action is distinct from its melting activity of liquid droplets. Alcohols, such as 1,6-HD, appear to remove water molecules around chromatin and locally condense chromatin. Therefore, liquid droplet results obtained using 1,6-HD should be carefully interpreted or reconsidered when these droplets are associated with chromatin.
This work was supported by JSPS and MEXT KAKENHI grants (19K23735, 20J00572, 18K06187, 19H05273 and 20H05936), a Japan Science and Technology Agency CREST grant (JPMJCR15G2), Takeda Science Foundation, Uehara Memorial Foundation, NIG Postdoctoral Fellowship and JSPS Postdoctoral Fellowship (PD).
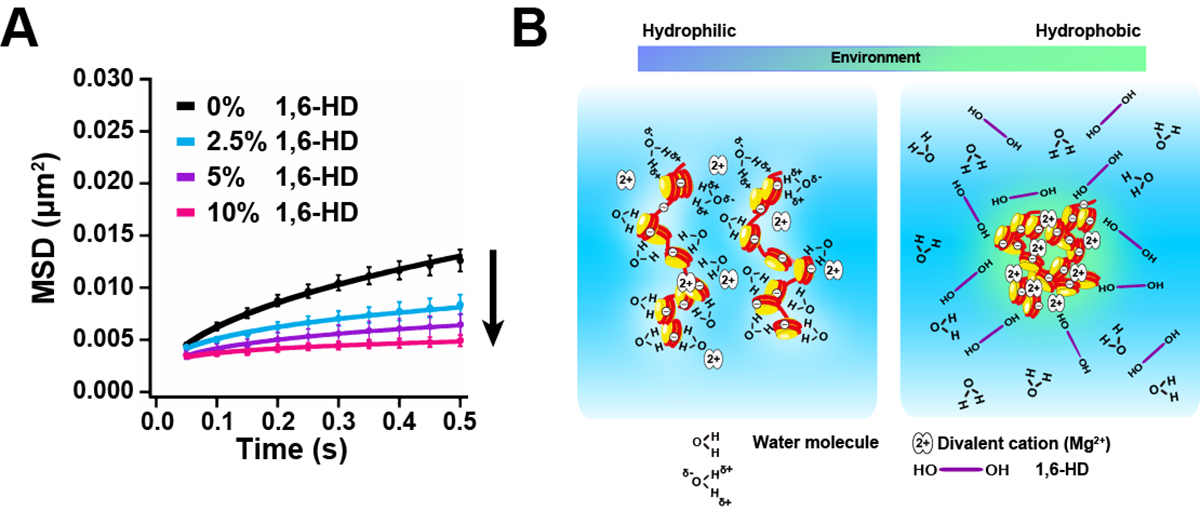
Figure: Chromatin immobilization by 1,6-HD and scheme for chromatin condensation.
(A) Mean square displacement plots (±SD among cells) of nucleosome motion in HeLa cells treated with 2.5% (light blue), 5% (purple), or 10% (pink) of 1,6-HD for 5–30 min. (B)(Left) Chromatin is associated with many water molecules with electrostatic interactions. (Right) Alcohols such as 1,6-HD can remove water molecules around chromatin, and its environment becomes more hydrophobic. This environmental change facilitates the formation of chromatin condensates. Note, this scheme is highly simplified and the molecules shown are not to scale. How 1,6-HD acts on chromatin at molecular level remains unclear.
The NIG Webinar: AID2 enables rapid target protein degradation in living mammalian cells and mice
The NIG international webinar by Prof. Kanemaki will be held on February 2nd at 4 pm, Eastern Time (on February 3rd at 6:00 am, Japan Standard Time). He will talk about a new genetic tool that enables rapid and reversible depletion of target proteins in living cells and animals. A Zoom link of the webinar will be obtained by free registration at the following URL (https://rois.zoom.us/meeting/register/tJAudOuprD4oHdV4dRlS6mAIdKX2F-jlAIEJ).
Time and Date:
Eastern Standard Time (EST): 4:00 pm, February 2nd
Pacific Standard Time (PST): 1:00 pm, February 2nd
Greenwich Mean Time (GMT): 9:00 pm, February 2nd
Central European Time (CET): 10:00 pm, February 2nd
Japan Standard Time (JST): 6:00 am, February 3rd
Registration (Zoom URL will be obtained by the free registration):
https://rois.zoom.us/meeting/register/tJAudOuprD4oHdV4dRlS6mAIdKX2F-jlAIEJ
Title:
AID2 enables rapid target protein degradation in living mammalian cells and mice
Speaker:
Prof. KANEMAKI, Masato
Molecular Cell Engineering Laboratory
National Institute of Genetics
Summary:
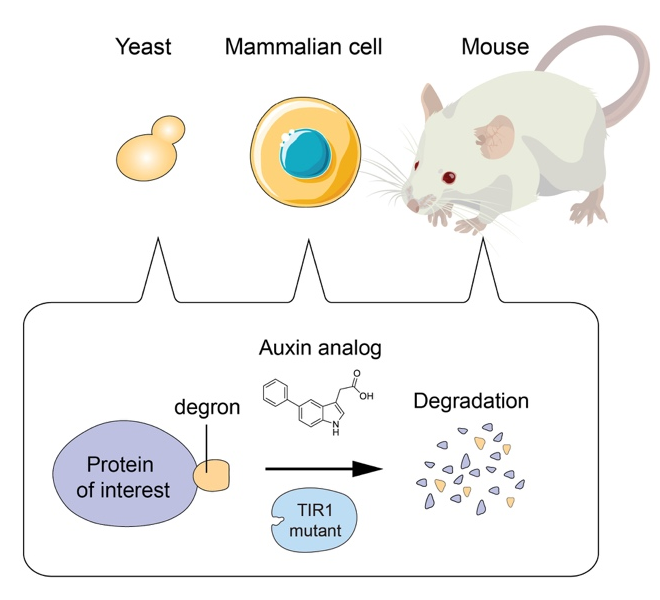
Genetic perturbation is a powerful way to analyze the function of proteins in living cells. For this purpose, we pioneered to develop the auxin-inducible degron (AID) technology by which a degron-fused protein can be rapidly degraded after the addition of the plant hormone auxin (Nishimura et al., Nat. Methods, 2009). By combining with CRISPR-based genome editing, it was possible to generate AID conditional mutants of human cells (Natsume et al., Cell Reports, 2016). The AID system became one of the popular genetic tools to study the function of proteins. However, leaky degradation and high doses of auxin for inducing degradation have been major drawbacks. Moreover, nobody has successfully applied the AID system to control protein degradation in living mice. We recently overcame these problems by taking advantage of chemical biology and successfully established the AID2 system (Yesbolatova et al. Nature Communications, 2020). By using AID2, we can now sharply control protein degradation in yeast, mammalian cells and mice.
• Link to KANEMAKI laboratory
• EurekAlert! link about the paper, Yesbolatova et al. Nature Communications, 2020
• Link to an interview of Prof. KANEMAKI
Chairperson: MAESHIMA, Kazuhiro















