Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Recovered: About the error that seminar information is not displayed correctly
The problem that seminar information was not displayed correctly in some browser versions has been fixed.
Thank you for your understanding.
Fluid-like behavior of chromatin in living human cell
Press release
Organization of fast and slow chromatin revealed by single-nucleosome dynamics
S. S. Ashwin, Tadasu Nozaki, Kazuhiro Maeshima, and Masaki Sasai
PNAS first published September 16, 2019 DOI:10.1073/pnas.1907342116
Press release (In Japanese only)
Understanding chromatin organization and dynamics is important, since they crucially affect DNA functions. In this study, we investigate chromatin dynamics by statistically analyzing single-nucleosome movement in living human cells. Bimodal nature of the mean square displacement distribution of nucleosomes allows for a natural categorization of the nucleosomes as fast and slow. Analyses of the nucleosome–nucleosome correlation functions within these categories along with the density of vibrational modes show that the nucleosomes form dynamically correlated fluid regions (i.e., dynamic domains of fast and slow nucleosomes). Perturbed nucleosome dynamics by global histone acetylation or cohesin inactivation indicate that nucleosome–nucleosome interactions along with tethering of chromatin chains organize nucleosomes into fast and slow dynamic domains. A simple polymer model is introduced, which shows the consistency of this dynamic domain picture. Statistical analyses of single-nucleosome movement provide rich information on how chromatin is dynamically organized in a fluid manner in living cells.
This research was supported by JST CREST(JPMJCR15G2), JSPS Kakenhi (JP19H05258, JP19H05273, JP19H01860, JP16H04746) and Takeda Science Foundation, RIKEN Pioneering Project、NIG Joint (2016-A2(6))
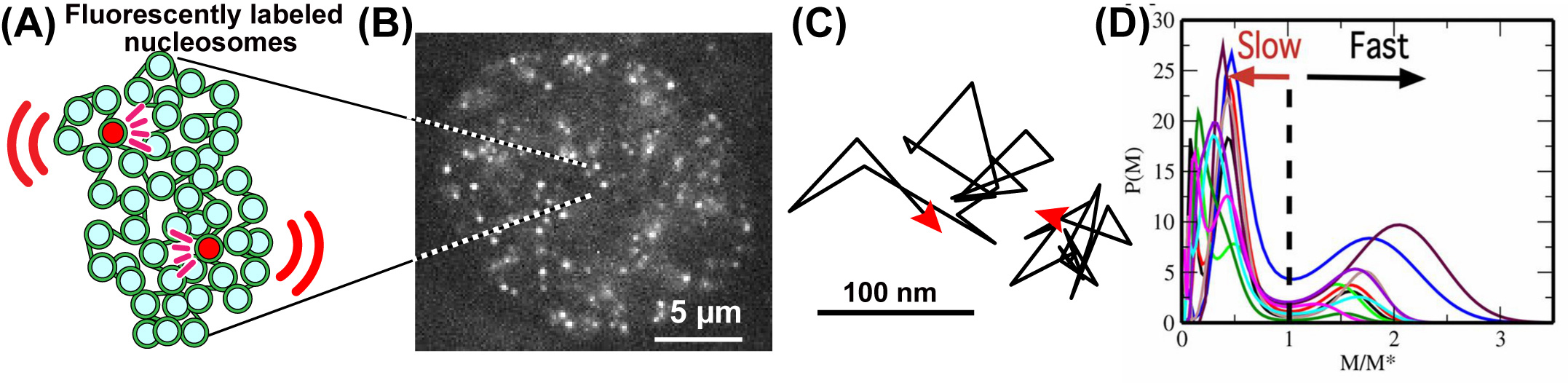
Fig: (A) A small fraction of nucleosomes, where DNA is wrapped around histone proteins, was fluorescently labeled (red). The labeled nucleosome movements can be tracked at super-resolution. (B) A single-nucleosome image of a living HeLa cell. (C) Representative two trajectories of the tracked single nucleosomes. (D) The distribution of MSD of single nucleosomes is plotted for 10-cell samples as functions of M/M*, where M* is M at the minimum between 2 peaks of the distribution.
Video: Raw video of single nucleosomes in the living HeLa cell. From Nozaki et al., (2017) Molecular Cell.
A new homologous recombination factor, HROB, controls the MCM8–MCM9 pathway
Control of homologous recombination by the HROB–MCM8–MCM9 pathway
Nicole Hustedt, Yuichiro Saito, Michal Zimmermann, Alejandro Álvarez-Quilón, Dheva Setiaputra, Salomé Adam, Andrea McEwan, Jing Yi Yuan, Michele Olivieri, Yichao Zhao, Masato T. Kanemaki, Andrea Jurisicova, and Daniel Durocher
Genes & Development online advanced publication DOI:10.1101/gad.329508.119
Most organisms including humans have two copies of genomic DNA, each of which is originated from the parents. Homologous Recombination (HR) is a reaction, which copy the sequence of donor DNA and paste it to the recipient DNA. HR plays a critical role in meiosis for gametogenesis and in damaged DNA repair during DNA replication or after irradiation, the latter of which is important to prevent cell death and cancer formation.
HR searches the donor sequence and synthesize DNA to copy and paste it to the recipient ones. Despite having a good understanding of the protein roles in the earlier steps in HR, much less is known about HR-associated DNA synthesis. We have studied MCM8–9, which is essential for HR-associated DNA synthesis. As a collaboration with the group led by Prof. Daniel Durocher at University of Toronto, we identified a new factor, HROB, which has an important role for loading MCM8–9 at the damaged DNA sites. Similar to the phenotypes of MCM8–9 loss, human cells lacking HROB showed a strong defect in DNA repair during DNA replication and HROB knockout mice were sterile, showing that, together with MCM8–9, HROB is important for genome maintenance and meiosis. We expect that elucidating HR, in which HROB and MCM8–9 are involved, would help cancer and fertility treatments in the future.
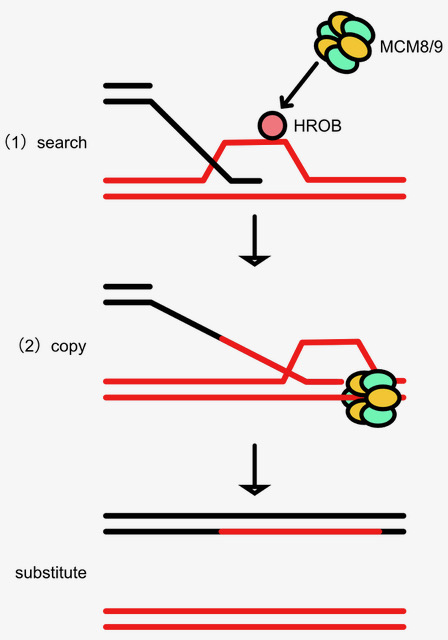
Figure: HR searches the donor sequence and uses DNA synthesis to copy and paste it to the recipient ones. In this paper, we found a novel factor, HROB, which promotes MCM8–9 loading to promote HR-associated DNA synthesis.
Message From NIGINTERN 2019
Neural signatures of sleep in zebrafish
Neural signatures of sleep in zebrafish.
Louis C Leung, Gordon X Wang, Romain Madelaine, Gemini Skariah, Koichi Kawakami, Karl Deisseroth, Alexander E Urban, and Philippe Mourrain
Nature 571(7764) 198-204 (2019) DOI:10.1038/s41586-019-1336-7
Slow-wave sleep and rapid eye movement (or paradoxical) sleep have been found in mammals, birds and lizards, but it is unclear whether these neuronal signatures are found in non-amniotic vertebrates. Here we develop non-invasive fluorescence-based polysomnography for zebrafish, and show—using unbiased, brain-wide activity recording coupled with assessment of eye movement, muscle dynamics and heart rate—that there are at least two major sleep signatures in zebrafish. These signatures, which we term slow bursting sleep and propagating wave sleep, share commonalities with those of slow-wave sleep and paradoxical or rapid eye movement sleep, respectively. Further, we find that melanin- concentrating hormone signalling (which is involved in mammalian sleep) also regulates propagating wave sleep signatures and the overall amount of sleep in zebrafish, probably via activation of ependymal cells. These observations suggest that common neural signatures of sleep may have emerged in the vertebrate brain over 450 million years ago.
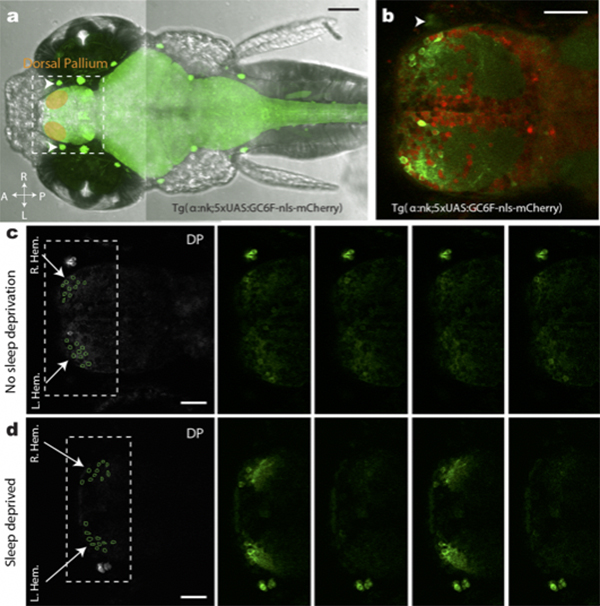
Figure: (a,b)Transgenic fish used for calcium imaging (c) Sleep non-deprived fish (d) slow bursting sleep seen in sleep deprived fish
▶This study is based on the previous study.















