DFAST: Prokaryotic genome annotation pipeline for data submission to DDBJ
DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication
Yasuhiro Tanizawa, Takatomo Fujisawa, Yasukazu Nakamura Bioinformatics 34(6) 1037-9, 2018 DOI:10.1093/bioinformatics/btx713Current technology of high throughput sequencing forces microbiologists to deal with more and more genomic data than before. In addition, annotated genome data must be deposited in the public sequence database; however, its complicated process is an another burden for researchers. We have developed an automatic prokaryotic genome annotation pipeline called DDBJ Fast Annotation and Submission Tool (DFAST) aiming to assist data submission to DDBJ. DFAST is available as a web tool, which allows users to create data submission files using graphical user interface, and also as a stand-alone tool, which is well suited for massive data processing on a local machine. DFAST can annotate a typical-sized bacterial genome within 5 minutes and has unique features such as pseudogene detection. It is available at https://dfast.ddbj.nig.ac.jp.
DFAST will facilitate the use of genomic data in microbial studies.
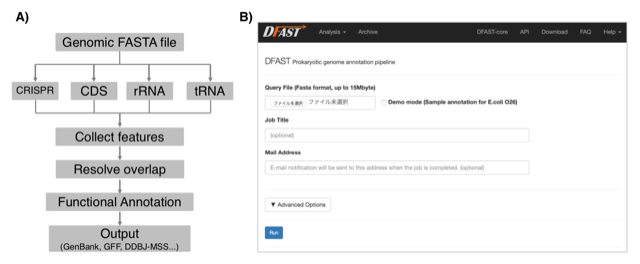
Figure: A) Annotation workflow of DFAST. B) Job submission form of the DFAST web server.















