Dynamic chromatin organization
Dynamic chromatin organization without the 30-nm fiber
Kazuhiro Maeshima, Satoru Ide and Michael Babokhov
Current Opinion in Cell Biology Volume 58, June 2019, Pages 95-104 DOI:10.1016/j.ceb.2019.02.003
Chromatin in eukaryotic cells is a negatively charged polymer composed of DNA, histones, and various associated proteins. Over the past ten years, our view of chromatin has shifted from a static regular structure to a dynamic and highly variable configuration. While the details are not fully understood yet, chromatin forms numerous compact domains that act as dynamic functional units of the genome in higher eukaryotes. By altering DNA accessibility, the dynamic nature of chromatin governs various genome functions including RNA transcription, DNA replication, and DNA repair/recombination. Based on new evidence coming from both genomics and imaging studies, we discuss the structural and dynamic aspects of chromatin and their biological relevance in the living cell.
This research was supported by JST CREST(JPMJCR15G2), JSPS Kakenhi (16H04746) and Takeda Science Foundation.
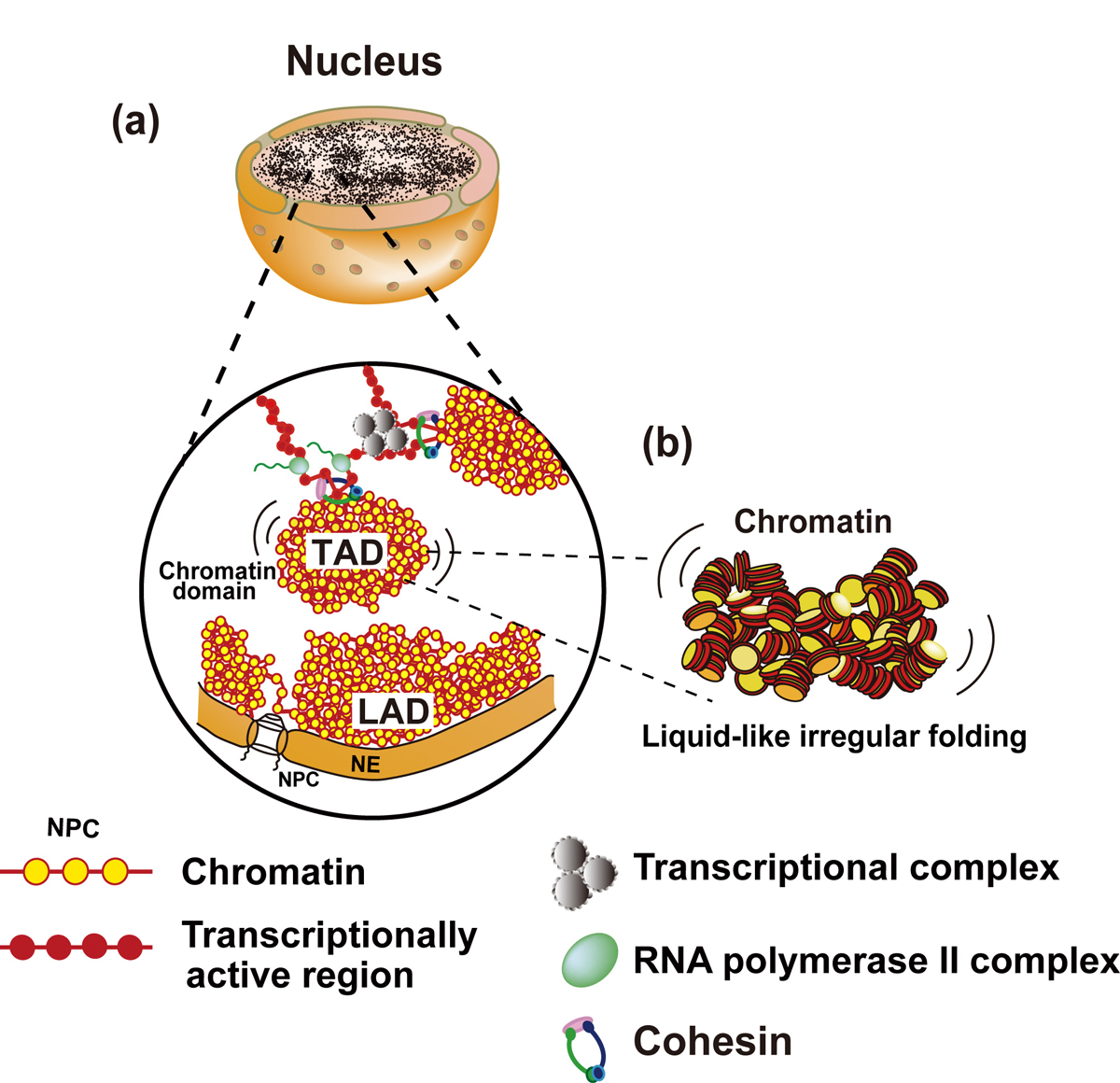
Figure: Interphase chromatin structure
(a and b) Chromatin consists of irregularly folded 10 nm fibers and forms numerous chromatin domains (e.g., topologically associating domains or contact/loop domains). The domains are formed by cohesin and nucleosome-nucleosome interactions. The compact domains could behave similar to a “liquid drop”. Binding of large transcriptional complexes (grey spheres) and RNA polymerase II (green spheres) might constrain movements of chromatin domains. Chromatin regions attached to nuclear envelop (NE) are called LADs (lamina-associated domains). NPC, nuclear pore complex; NE, nuclear envelop.















