Transcription regulates dynamic movements of genomic DNA in living human cells
Press release
Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II
Ryosuke Nagashima, Kayo Hibino, S. S. Ashwin, Michael Babokhov, Shin Fujishiro, Ryosuke Imai, Tadasu Nozaki, Sachiko Tamura, Tomomi Tani, Hiroshi Kimura, Michael Shribak, Masato T.Kanemaki, Masaki Sasai, and Kazuhiro Maeshima
Journal of Cell Biology Published March 1, 2019 DOI:10.1083/jcb.201811090
Press release (In Japanese only)
The human body is composed of over forty trillion cells. Within each of these cells there is close to two meters of tightly packaged genomic DNA, the blueprint of life. Recently, there have been many advances in understanding how DNA is packaged and organized in the cell. In contrast, how DNA behaves mostly remains unknown.
In this recently published report, SOKENDAI graduate student Ryosuke Nagashima, assistant professor Kayo Hibino and professor Kazuhiro Maeshima of the National Institute of Genetics partnered with assistant professor S.S. Ashwin and professor Masaki Sasai of Nagoya University to study the movements of DNA in living cells using super resolution fluorescence microscopy (Figure 1A and B). Previously, it was commonly thought that the process of reading genomic information, known as transcription, would lead to more dynamic movements of DNA through the loosening of nucleosome packing. However, as Nagashima et al. report, transcription normally restricts DNA movement since the inhibition of transcription leads to increased DNA movements (Figure 1C). Furthermore, this work revealed that RNA polymerase II and other transcription-related factors form hubs to limit the motion of DNA (Figure 2). These results suggest that hubs may network the genome, restricting DNA movement to enable the efficient execution of transcription.
The results of this research have advanced our understanding as to how genetic information is acquired from DNA and may provide further clues concerning related disease states caused by abnormal changes to transcription.
This research is a result of a collaboration between the Genome Dynamics Laboratory of the National Institute of Genetics (Ryosuke Nagashima, Kayo Hibino, Michael Babokhov, Sachiko Tamura, Tadasu Nozaki, Ryosuke Imai, Kazuhiro Nagashima), the Nagoya University Department of Computational Science and Engineering (S.S. Ashwin, Shin Fujishiro, Masaki Sasai), the Cell Biology Center, Institute of Innovative Research, Tokyo Institute of Technology (Hiroshi Kimura), the Division of Molecular Cell Engineering of the National Institute of Genetics (Masato T. Kanemaki) and the Eugene Bell Center for Regenerative Biology and Tissue Engineering, Marine Biological Laboratory, Woods Hole (Michael Shribak, Tomomi Tani).
This research was supported by JST CREST(JPMJCR15G2), JSPS Kakenhi(16H04746), Takeda Science Foundation, RIKEN Pioneering Project, NIG-JOINT(2016-A2 (6)), SOKENDAI and National Institute of General Medical Sciences grant (R01-GM101701).
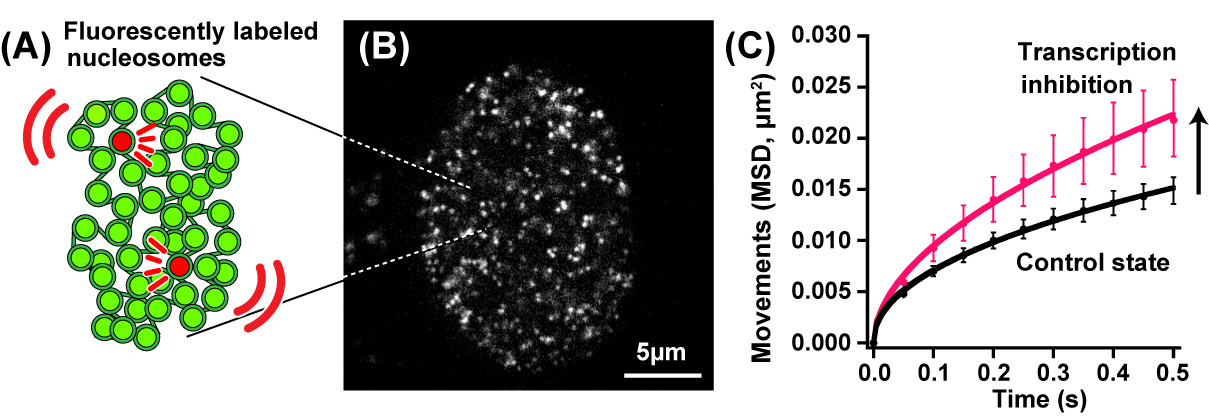
Fig.1. (A) A small fraction of nucleosomes was fluorescently labeled (red). (B) A single-nucleosomes image of a living human cell. (C) Genomic DNA motion, which is shown as mean square displacement, MSD), increased upon transcription inhibition.
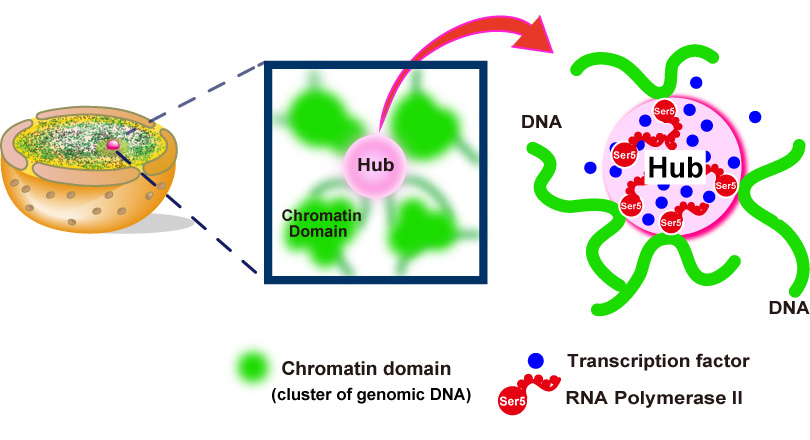
Fig.2. Hubs (pink spheres) of RNA polymerase II (red) and other components (blue spheres) of the gene transcription machinery constrain genomic DNA movements by connecting different genome regions together into an organized network.
Video1: Movie data (50 ms/frame) of single nucleosomes fluorescently labeled in a living human cell. Note that clear, well-separated dots and their movements were visualized.
Video2: Movie data (50 ms/frame) of single nucleosomes upon transcription inhibition. Note that greater movements of the dots are observed than those in Video1, suggesting the local genome dynamics increased upon transcription inhibition.















