Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Call for Urgent Joint Research with Researchers in Hokkaido Eastern Iburi Earthquake Disaster Area
A simple and efficient multiple sgRNA expression system in zebrafish
A tRNA-based multiplex sgRNA expression system in zebrafish and its application to generation of transgenic albino fish.
Tomoya Shiraki and Koichi Kawakami
Scientific Reports 8, Article number: 13366 (2018) DOI:10.1038/s41598-018-31476-5
The CRISPR/Cas9 system can be introduced into zebrafish fertilized eggs by microinjection and facilitates gene knock-out, genome editing, and gene knock-in studies in zebrafish. Further, expression of gRNA and controlled expression of Cas9 in transgenic fish have enabled the study of gene functions in specific cell types in zebrafish. The transgenic Cas9 approach would be more useful if multiple sgRNAs could be expressed simultaneously since we could knock-out a gene more efficiently or disrupt multiple genes simultaneously. However, such a system had not been developed in zebrafish.
In this study, we describe a novel system to express multiple sgRNAs efficiently in zebrafish, that relies on the endogenous tRNA processing machinery. In this study, we demonstrated that active sgRNAs can be produced from tRNA-fused precursor transcripts. To show a proof of principle, we constructed a transgenic fish line expressing a single transcript containing three distinct sgRNAs, that targeted the slc45a2 (albino) gene. We found that the transgenic-albino fish showed nearly complete albino phenotypes, which were transparent enough for imaging at the cellular resolution to be performed. Thus, the tRNA-based multiplex sgRNA expression system should facilitate genetic studies of gene functions in zebrafish.
This study was supported by JSPS KAKENHI Grant Numbers (JP15H02370, JP16K20983, JP16H01651, JP18H04988), and NBRP and NBRP/Genome Information upgrading program from Japan Agency for Medical Research and Development (AMED).
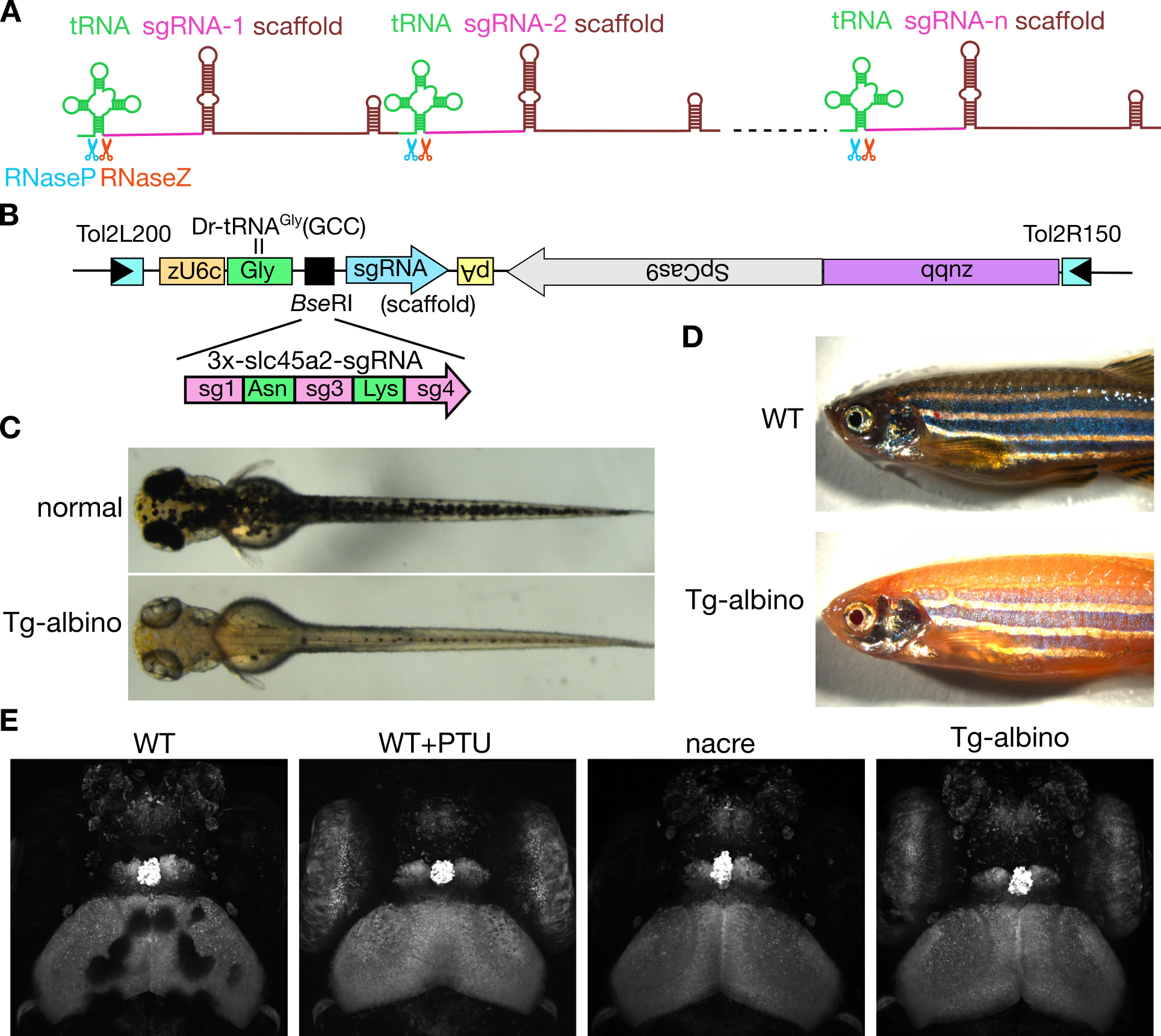
Figure: (A) Schematic diagram of the polycistronic tRNA-sgRNA system for multiplex genome editing. (B) DNA construct used for the generation of transgenic albino (Tg-albino) zebrafish. Three distinct sgRNAs targeting the albino (slc45a2) locus were expressed. (C-E) The Tg-albino fish showed nearly complete albino phenotypes, which were transparent enough for imaging at the cellular resolution. (C) Tg-albino at 3-days post-fertilization (dpf). (D)Adult Tg-albino fish. (E) Fluorescent images of a GFP-transgenic fish at 5 dpf in Tg-albino background.
Differential dynamics of cortical neuron dendritic trees revealed by long-term in vivo imaging in neonates
![]()
Differential dynamics of cortical neuron dendritic trees revealed by long-term in vivo imaging in neonates
Shingo Nakazawa, Hidenobu Mizuno, Takuji Iwasato
Nature Communications 9, Article number: 3106 (2018) DOI:10.1038/s41467-018-05563-0
Pressrelease (In Japanese only)
Proper neuronal circuit function relies on precise dendritic projection, which is established through activity-dependent refinement during early postnatal development. Here we revealed dynamics of dendritic refinement in the mammalian brain by conducting long-term imaging of the neonatal mouse barrel cortex. By “retrospective” analyses, we identified “prospective” barrel-edge spiny stellate (SS) neurons in early neonates, which had an apical dendrite and primitive basal dendrites (BDs). These neurons retracted the apical dendrite gradually and established strong BD orientation bias through continuous “dendritic tree” turnover. A greater chance of survival was given to BD trees emerged in the barrel-center side, where thalamocortical axons (TCAs) cluster. When the spatial bias of TCA inputs to SS neurons was lost, BD tree turnover was suppressed, and most BD trees became stable and elaborated mildly. Thus, barrel-edge SS neurons could establish the characteristic BD projection pattern through differential dynamics of dendritic trees induced by spatially biased inputs.
Source: Nature Communications 9, Article number: 3106 (2018) DOI:10.1038/s41467-018-05563-0
Genetic factors contributing to ‘strabismus’ — or misaligned eyes
![]()
Protocadherin-mediated cell repulsion controls the central topography and efferent projections of the abducens nucleus
Kazuhide Asakawa, Koichi Kawakami
Cell Reports Volume 24, ISSUE 6, P1562-1572, August 07, 201 DOI:10.1016/j.celrep.2018.07.024
Press release (In Japanese only)
Approximately 2-3 % of children suffer from a condition called strabismus, which is more commonly known as “lazy eye” or “crossed eyes”. The majority of cases are thought to involve both genetic and environmental factors. Knowing the genetic causes of strabismus is very important as it would help estimate risk of developing the condition and increase the chances of initiating early treatment to prevent impaired eye movement.
A major obstacle to uncovering the genetic causes of strabismus has been the physical limitation of observing the connections between the brain and eye muscles. These are formed by long cables called “axons” that are extended from brain cells deep inside the head at a very early developmental stage. Therefore it is difficult to observe the fine architecture of the brain-eye muscle connections at the cellular level and analyze how they might be affected by gene mutations.
To overcome this difficulty, researchers at the National Institute of Genetics in Japan chose as their experimental model a tropical fish called zebrafish. Zebrafish larvae are nearly transparent while they are developing, so the researchers hoped that they could make the brain-eye muscle connections visible using genetic engineering. Employing their own original techniques, the team successfully created a zebrafish in which the specific group of brain cells that send signals for outward eye movement, called abducens motor neurons, as well as the targeted eye muscles emit differently colored fluorescent light clearly visible through the developing fish’s transparent head.
These special zebrafish proved instrumental in allowing the team to make great progress in uncovering the genetic mechanism of strabismus. The team demonstrated that the abducens motor neurons developed abnormally when the gene called protocadherin 17 (pcdh17) was mutated. This gene encodes for the production of the Pcdh17 protein that is located on the cell surface of the abducens motor neurons. When an abnormal Pcdh17 protein is produced from the manipulated pcdh17 gene and located on the cell surface, the neurons did not position properly in the brain and the axons they extended failed to reach the target eye muscle. In many cases these disabled neurons clumped together forming closely-packed cellular aggregates. These results suggest that an abducens motor neuron uses repulsive forces to both position itself correctly in the brain and extend its axon correctly to the target muscle and that these forces are mediated by the Pcdh17 protein molecules on its surface interacting with itself and/or other neurons around it. This study was published in Cell Reports on 7th August.
Dr. Asakawa, who led this research project, says, “Pcdh17 creates a moderate repulsive force between abducens motor neurons presumably allowing them to behave like a fluid forming a stream and flowing into the muscle. Without them, the neurons just become frozen like ice. Since Pcdh17 protein is also present in humans, it is likely to play a similar role in our body for the development of normal eye movement. Moreover, given that many types of brain neurons connecting with the head muscles are covered with different types of proteins that are similar to Pcdh17 on their cell surface, genetic mutations in these genes might increase the risk of developing other congenital disorders of craniofacial movements”.
Asakawa continues, “Compared to the connections between the spinal cord and skeletal muscles that generate our body movements, the brain and eye muscle connections have some special characteristics making them selectively resistant to degeneration in fatal motor neuron diseases, such as amyotrophic lateral sclerosis (ALS). We expect that the visual and genetically-manipulatable brain-eye muscle connection in fish also has great potential to reveal a way to protect motor neurons from degeneration in ALS.”
This study was supported by KAKENHI (JP15H02370, JP18H04988, JP22700349, JP23115720, and research grants from The Uehara Memorial Foundation, The Kao Foundation for Arts and Sciences, The Mitsubishi Foundation, Daiichi-Sankyo Foundation of Life Science and National BioResouce Project from Japan Agency for Medical Research and Development (AMED).
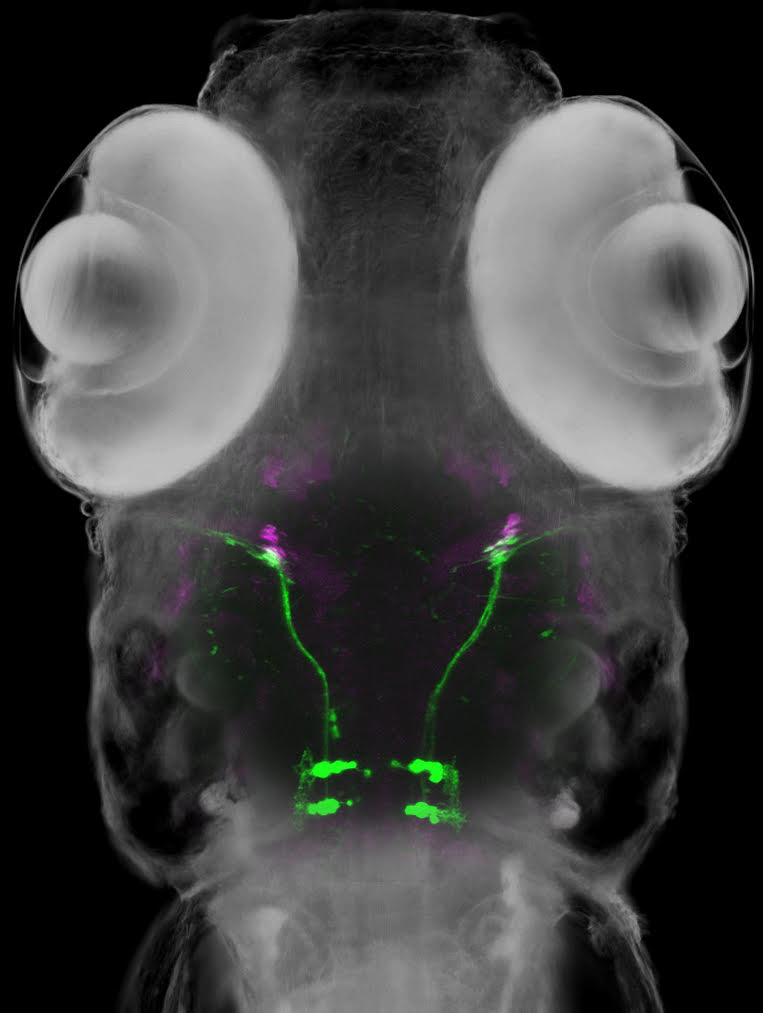
Figure1: A zebrafish larva expressing a green fluorescent protein in the abducens motor neurons and a red fluorescent protein in the eye muscle (magenta).
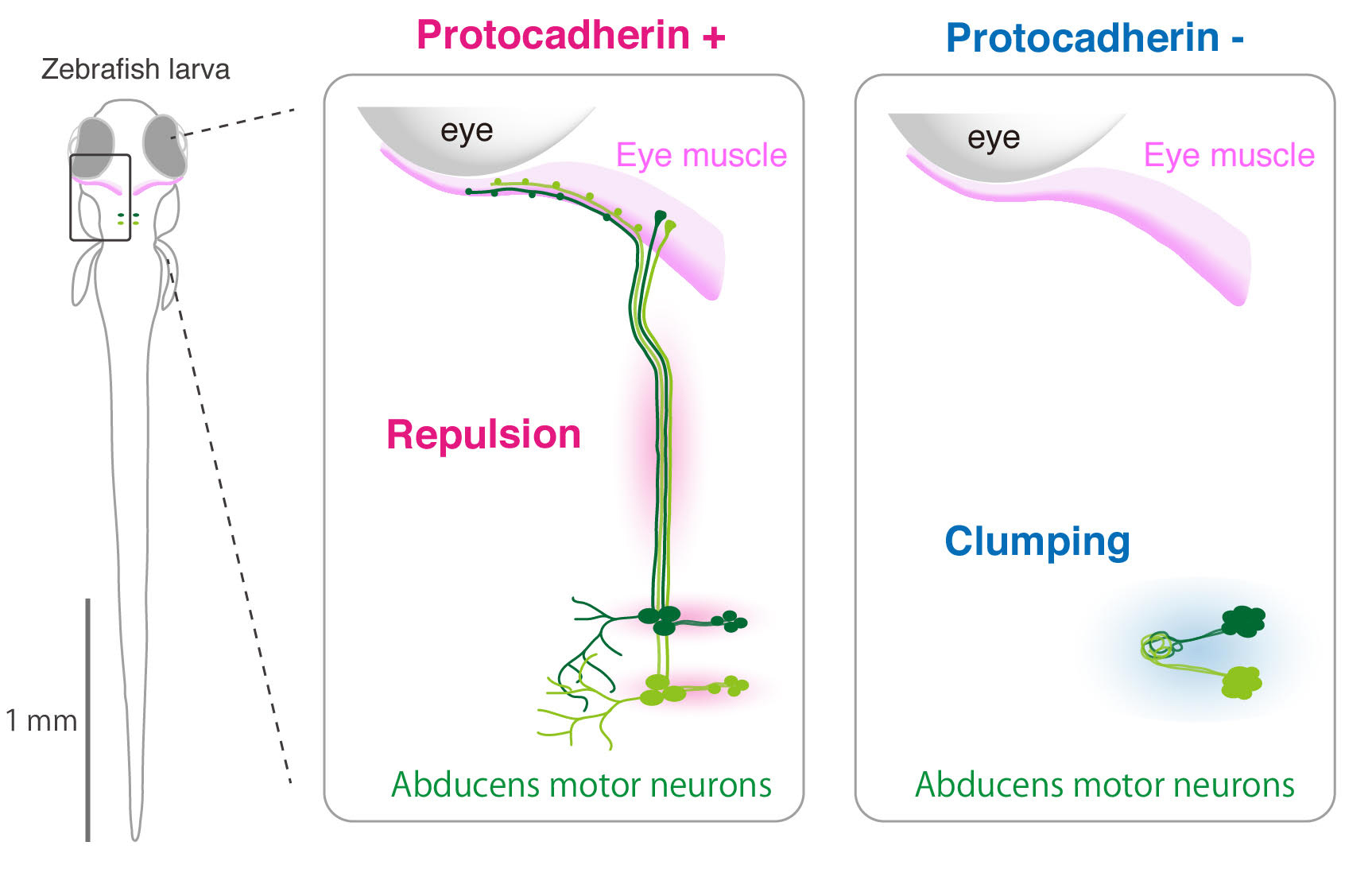
Figure2: Repulsive forces-mediated by protocadherins promote cell migration and axon ourgrowth of abducens motor neurons (left) . Loss of the rupulsive forces leads to clumping phenotype (right).
EurekAlert!, the online, global news service operated by AAAS, the science society, PUBLIC RELEASE: 7-AUG-2018
New assistant professor joins NIG
New assistant professor joins NIG as of September 1, 2018.















