An asymmetric attraction model for the diversity and robustness of cell arrangement in nematodes
Press release
An asymmetric attraction model for the diversity and robustness of cell arrangement in nematodes
Kazunori Yamamoto, Akatsuki Kimura
Development 2017 144: 4437-4449; DOI:10.1242/dev.154609
Pressrelease (In Japanese only)
During early embryogenesis in animals, cells are arranged into a species-specific pattern in a robust manner. Diverse cell arrangement patterns are observed, even among close relatives (Fig. A). In the present study, we evaluated the mechanisms by which the diversity and robustness of cell arrangements are achieved in developing embryos. We successfully reproduced various patterns of cell arrangements observed in various nematode species in Caenorhabditis elegans embryos by altering the eggshell shapes (Fig. B). The findings suggest that the observed diversity of cell arrangements can be explained by differences in the eggshell shape. Additionally, we found that the cell arrangement was robust against eggshell deformation. Computational modeling revealed that, in addition to repulsive forces, attractive forces are sufficient to achieve such robustness (Fig. C). The present model is also capable of simulating the effect of changing cell division orientation. Genetic perturbation experiments demonstrated that attractive forces derived from cell adhesion are necessary for the robustness. The proposed model accounts for both diversity and robustness of cell arrangements, and contributes to our understanding of how the diversity and robustness of cell arrangements are achieved in developing embryos.
This research article was published in the special issue of Development ‘On Growth and Form – 100 years on’, inspired by the seminal work of D’Arcy Thomson. See the authors’ comments:
Kazunori Yamamoto, the first author of the paper received Morishima Award for this research.
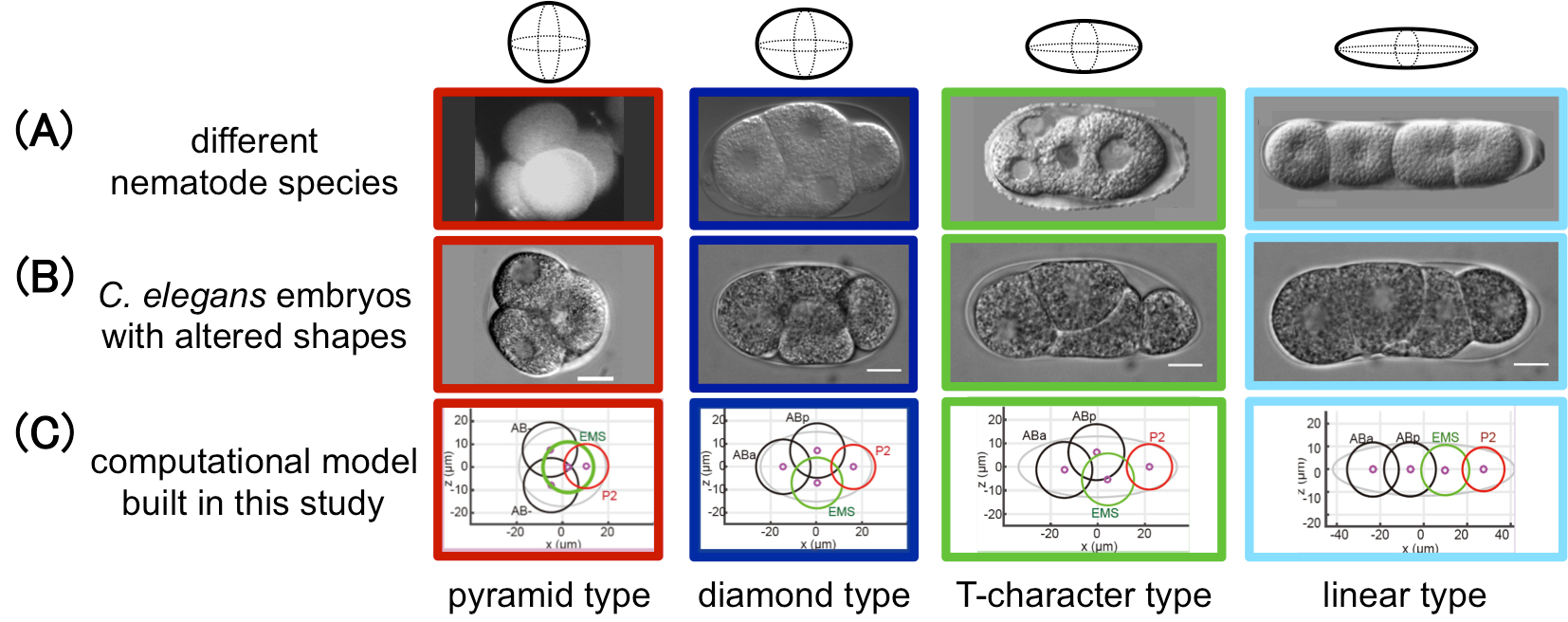
Figure: This study successfully reproduced the cell arrangement patterns of various nematode species (A) using C. elegans by altering the shape of the eggshell (B). A computational model was also built in this study that reproduces the diversity and robustness of the cell arrangement patters (C). In (A), the patterns of Enoplus brevis (red, image from Schulze and Shierenberg, Evodevo 2, 18, 2011), C. elegans (blue, this study), Cephalobus sp. (green, Goldstein, Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1521-1531, 2001), and Belonolaimus sp. (light blue, Goldstein, 2001) are shown.
Source: Development 2017 144: 4437-4449
DOI: 10.1242/dev.154609















