A new function of rDNAs to compact bacterial chromosome
Microbial Genetics Laboratory / Niki Group
Multiple cis-acting rDNAs contribute to nucleoid separation and recruit the bacterial condensin, Smc-ScpAB
Koichi Yano, and Hironori Niki
Cell Reports, Vol. 21 Iss. 5, October 31, 2017 DOI:http://dx.doi.org/10.1016/j.celrep.2017.10.014
Condensins load onto DNA to organize chromosome. Smc-ScpAB clearly loads onto the parS sites bound by Spo0J, but other loading site(s) must operate independently of parS. In this study, we asked where and how Smc-ScpAB normally selects its loading site. Our results suggest that rDNA is also a loading site. A pull-down assay revealed that Smc-ScpAB preferentially loads onto rDNA in the wild-type cell and even in a Δspo0J mutant, but not in a Δsmc mutant. Moreover, we showed that deletion mutants of rDNAs cause a defect in nucleoid separation, and at least two rDNAs near oriC are essential for separation. Full-length rDNA including promoters is required for loading and nucleoid separation. A synthetic defect by deletions of both rDNA and spo0J resulted in more aberrant nucleoid separation. We propose that a single-stranded segment DNA that is exposed at highly transcribed rRNA operons would become a target for Smc-ScpAB loading.
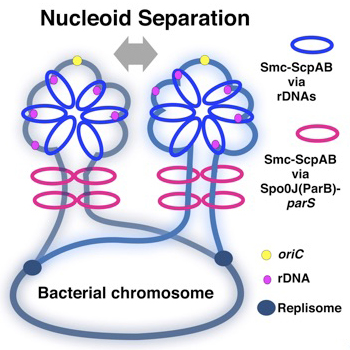
Schemes of compaction of newly replicated DNA strands. The yellow circle indicates oriC, black blue circles indicate replication forks, and magenta circles indicate rrn operons. The blue rings indicate Smc-ScpAB topologically loaded on rrn operons, and magenta rings represent Smc-ScpAB topologically loaded by ParB bound to parS as shown in Wang et al. (2017). In addition to separation of oriC, chromosome compaction would always be accompanied by proper chromosome segregation. The chromosomal region adjacent to oriC is packed by Smc in the WT so that the nucleoids are clearly separated.















