Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Spinal RacGAP α-chimaerin is required to establish the midline barrier for proper corticospinal axon guidance
Press release
Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis
Toyoaki Natsume, Kohei Nishimura, Sheroy Minocherhomji, Rahul Bhowmick, Ian D. Hickson, Masato T. Kanemaki
Genes & Development DOI:10.1101/gad.297663.117
Pressrelease (In Japanese only)
Dr. Toyoaki Natsume and Prof. Masato Kanemaki at National Institute of Genetics, ROIS, together with the group led by Prof. Ian D. Hickson at University of Copenhagen, reported a new system to deal with failure in DNA replication. The finding was published in Genes & Development in advance of the print journal.
In cell proliferation, genomic DNA has to be precisely copied into two (DNA replication) before equal distribution to two daughter cells. For doing this, double-stranded DNA is unwound, the process of which is similar to open a zipper of your clothes (Figure 1A). Similar to pulling the ‘slider’ for opening a zipper, the replicative helicase known as MCM2–7 moves on DNA for unwinding double-stranded DNA. However, because human genomic DNA is very long (approx. 2 m per cell), it is challenging to entirely unwind the genomic DNA. Occasionally, the MCM2–7 helicase falls off from DNA when it encounters a roadblock (such as DNA damage). Because reloading of MCM2–7 is strictly inhibited during S phase (when cells carry out DNA replication), this might lead to incompletion of DNA replication, which causes the loss of genetic information from daughter cells unless the cells have a mechanism to deal with the problem.
In this study, the research groups observed how human cells responded to artificial removal of the MCM2–7 helicase by using the auxin-inducible degron (AID) technology that they had developed previously (the information about this technology is described here). They revealed that the MCM8–9 helicase, which is evolutionally related to the MCM2–7 helicase, promotes a non-canonical DNA synthesis as a backup system after removal of MCM2–7 (Figure 1B).
Many anticancer drugs kills cancer cells by inducing DNA lesions, which enhance removal of MCM2–7 from DNA during DNA replication. An inhibitor of the MCM8–9 helicase might enhance the effect of existing anticancer drugs by shutting off this backup system (Figure 2).
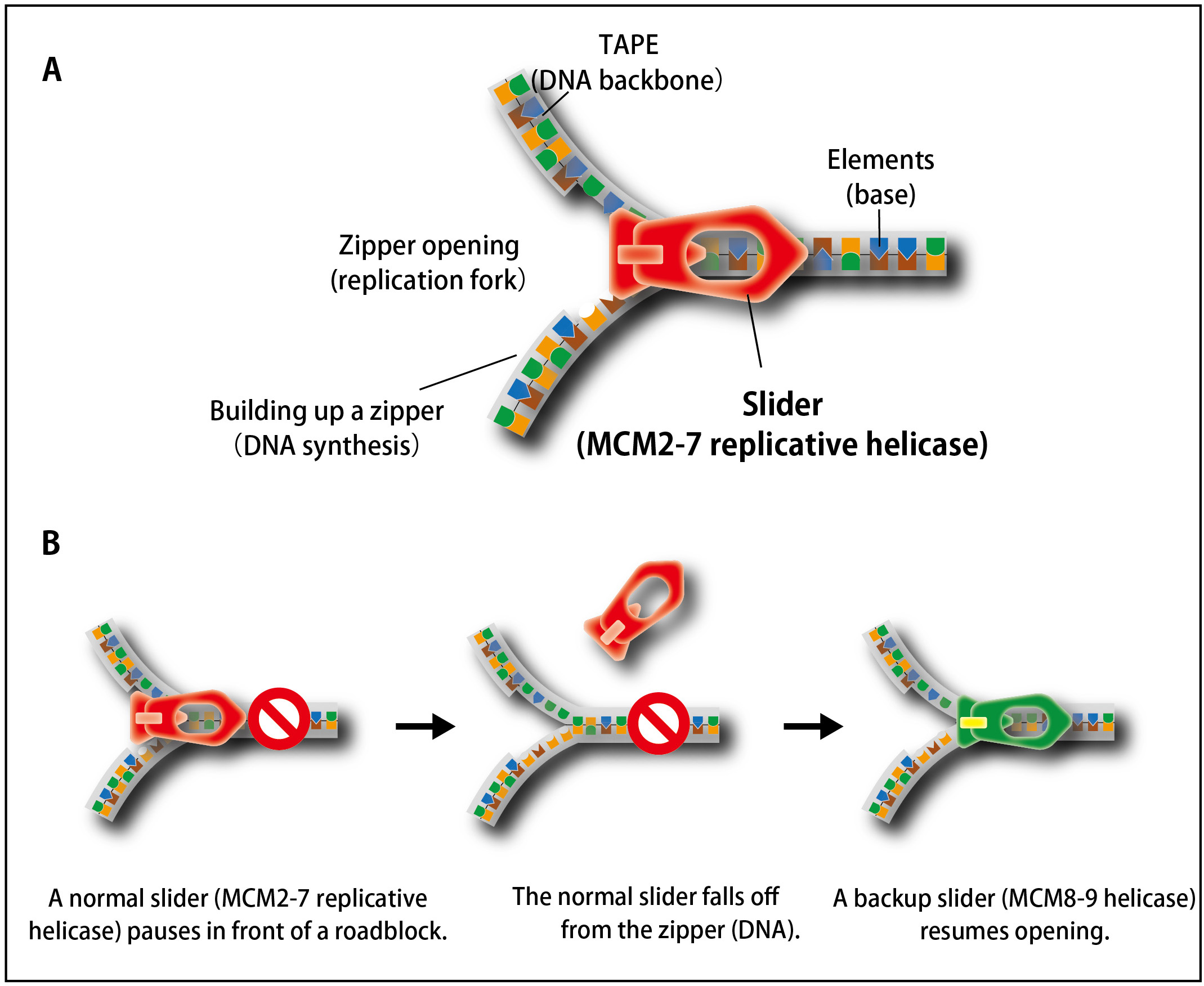
Figure 1. The process of unwinding double-stranded DNA during DNA replication is similar to that of opening a zipper of your clothes.
(A) Similar to the slider that opens a zipper, the MCM2–7 replicative helicase opens double-stranded DNA in cells.(B) When the replicative MCM2–7 helicase (a normal slider) encounters to a roadblock, it occasionally falls off from DNA. To continue DNA synthesis, cells recruit the MCM8–9 helicase as a backup slider.
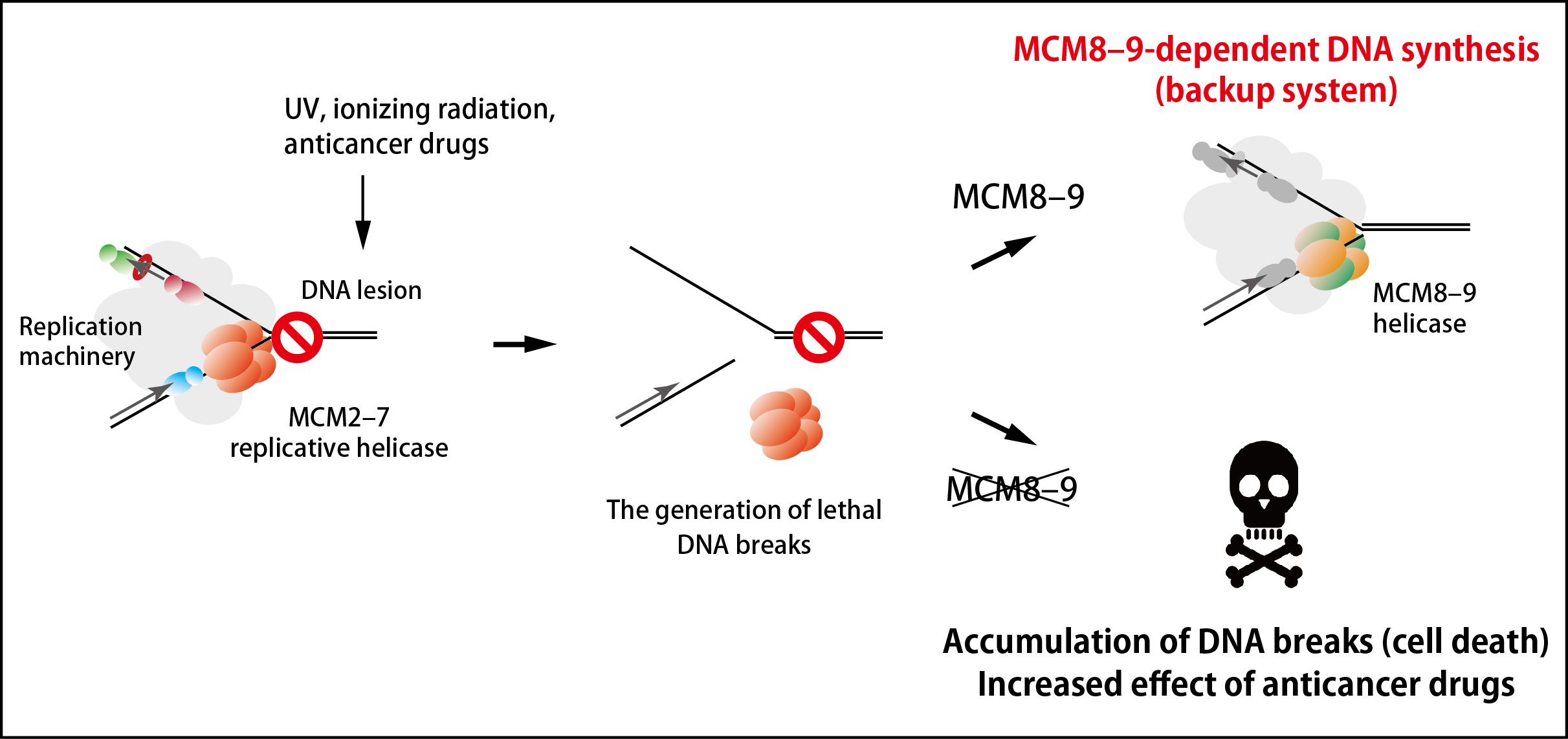
Figure 2. An MCM8–9-dependent backup system against failure in DNA replication.
The MCM2–7 replicative helicase falls off when encountered to a roadblock on DNA, leading to generation of DNA breaks. In this study, the research groups found that the MCM8–9 helicase continues DNA synthesis on behalf of the MCM2–7 (top right). If MCM8–9 does not work, the accumulation of DNA breaks results in cell death (bottom right).
Density imaging of chromatin in live cells
Biological Macromolecules Laboratory / Maeshima Group
Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy
Ryosuke Imai, Tadasu Nozaki, Tomomi Tani, Kazunari Kaizu, Kayo Hibino, Satoru Ide, Sachiko Tamura, Koichi Takahashi, Michael Shribak, and Kazuhiro Maeshima
Molecular Biology of the Cell, 2017 DOI:10.1091/mbc.E17-06-0359
In eukaryotic cells, highly condensed inactive/silenced chromatin has long been called “heterochromatin.” However, recent research suggests that such regions are in fact not fully transcriptionally silent, and that there exists only a moderate access barrier to heterochromatin. To further investigate this issue, it is critical to elucidate the physical properties of heterochromatin such as its total density in live cells. Here, using orientation-independent differential interference contrast (OI-DIC) microscopy, which is capable of mapping optical path differences, we investigated the density of the total materials in pericentric foci, a representative heterochromatin model, in live mouse NIH3T3 cells. We demonstrated that the total density of heterochromatin (208 mg/mL) was only 1.53-fold higher than that of the surrounding euchromatic regions (136 mg/mL) while the DNA density of heterochromatin was 5.5- to 7.5-fold higher. This surprisingly small difference may be due to that non-nucleosomal materials (proteins/RNAs)(∼120 mg/mL) are dominant in both chromatin regions. Monte Carlo simulation suggested that non-nucleosomal materials contribute to creating a moderate access barrier to heterochromatin, allowing minimal protein access to functional regions. Our OI-DIC imaging offers insight into the density of live cellular environments.
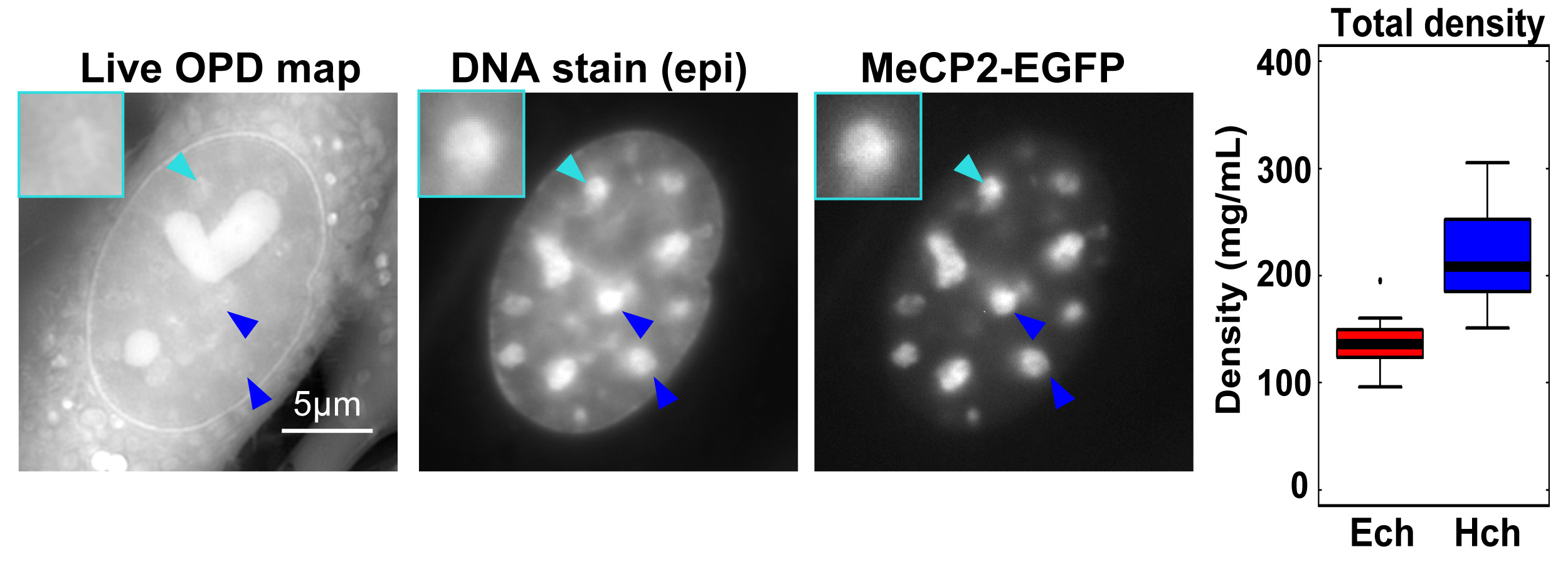
Typical images of the Optical Path Distance (OPD) map (reflecting density at each pixel) by OI-DIC, DNA staining, and MeCP2-EGFP (heterochromatin marker) signals in live NIH3T3 cells. Large foci seen in DNA staining and MeCP2-EGFP images (arrowheads) were assumed to be heterochromatin. Note that the OPD of the foci was similar or only slightly higher than that of the surrounding euchromatin. (Right) The analyzed total densities of pericentric heterochromatin foci (Hch, 208 mg/mL) and euchromatin (Ech, 136 mg/mL). The median density ratio between them was 1.53.
Summer Holiday (Aug. 14,15)
NIG will be closed from August 14 and August 15, 2017 for summer holiday.
Thank you for your understanding and cooperation.
Call for Collaboration Program at ROIS Joint Support-Center for Data Science Research (DS)
Call for Collaboration Program at ROIS Joint Support-Center for Data Science Research (DS)
Social hierarchy affects depression-like behavior and gene expression in mouse brain
Press release
Hierarchy in the home cage affects behaviour and gene expression in group-housed C57BL/6 male mice
Yasuyuki Horii, Tatsuhiro Nagasawa, Hiroyuki Sakakibara, Aki Takahashi, Akira Tanave, Yuki Matsumoto, Hiromichi Nagayama, Kazuto Yoshimi, Michiko T. Yasuda, Kayoko Shimoi, Tsuyoshi Koide
Scientific Reports Article number: 6991 (2017) DOI:10.1038/s41598-017-07233-5
Pressrelease (In Japanese only)
Social stress is one of the major causes of depression in humans. It is therefore important that the effects of social stress on behaviour and gene expression in the brain are studied. We performed experiments using male mice to analyse social hierarchy in groups of mice in cages and investigated emotional behaviour and hippocampal gene expression in the mice. We found significantly different emotional behaviour and expression of genes associated with the serotonergic system in dominant and subordinate mice. Treating the mice with a selective serotonin reuptake inhibitor antidepressant restored gene expression in subordinate mice and caused the emotional behaviour of the subordinate mice to be recovered, suggesting that alterations to the serotonergic system are key factors in phenotypic changes caused by the social ranks of subordinate mice. We believe that the results of this study are important because they improve our understanding of how social stress induces mental health problems in humans.















