Systematic identification of regulatory elements derived from human endogenous retroviruses
Division of Human Genetics / Inoue Group
Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses.
Jumpei Ito, Ryota Sugimoto, Hirofumi Nakaoka, Shiro Yamada, Tetsuaki Kimura, Takahide Hayano, and Ituro Inoue.
PLoS Genetics. Jul 12;13(7):e1006883. 2017. DOI:10.1371/journal.pgen.1006883
Human endogenous retroviruses (HERVs) have regulatory elements that possibly influence the transcription of host genes. We systematically identified these HERV regulatory elements (HERV-REs) based on publicly available datasets of ChIP-Seq. Overall, 794,972 HERV-REs were identified. Clustering analysis showed that HERVs can be grouped according to the TF binding patterns; HERV groups bounded by pluripotent TFs (e.g., SOX2, POU5F1, and NANOG), endoderm TFs (e.g., GATA4/6, SOX17, and FOXA1/2), hematopoietic TFs (e.g., SPI1, GATA1/2, and TAL1), and CTCF were identified. Regulatory elements of HERVs tended to locate nearby genes involved in immune responses, indicating that the regulatory elements play an important role in controlling the immune regulatory network. Finally, we constructed dbHERV-REs, an interactive database of HERV regulatory elements (http://herv-tfbs.com/). This study provides fundamental information in understanding the impact of HERVs on host transcription, and offers insights into the transcriptional modulation systems of HERVs.
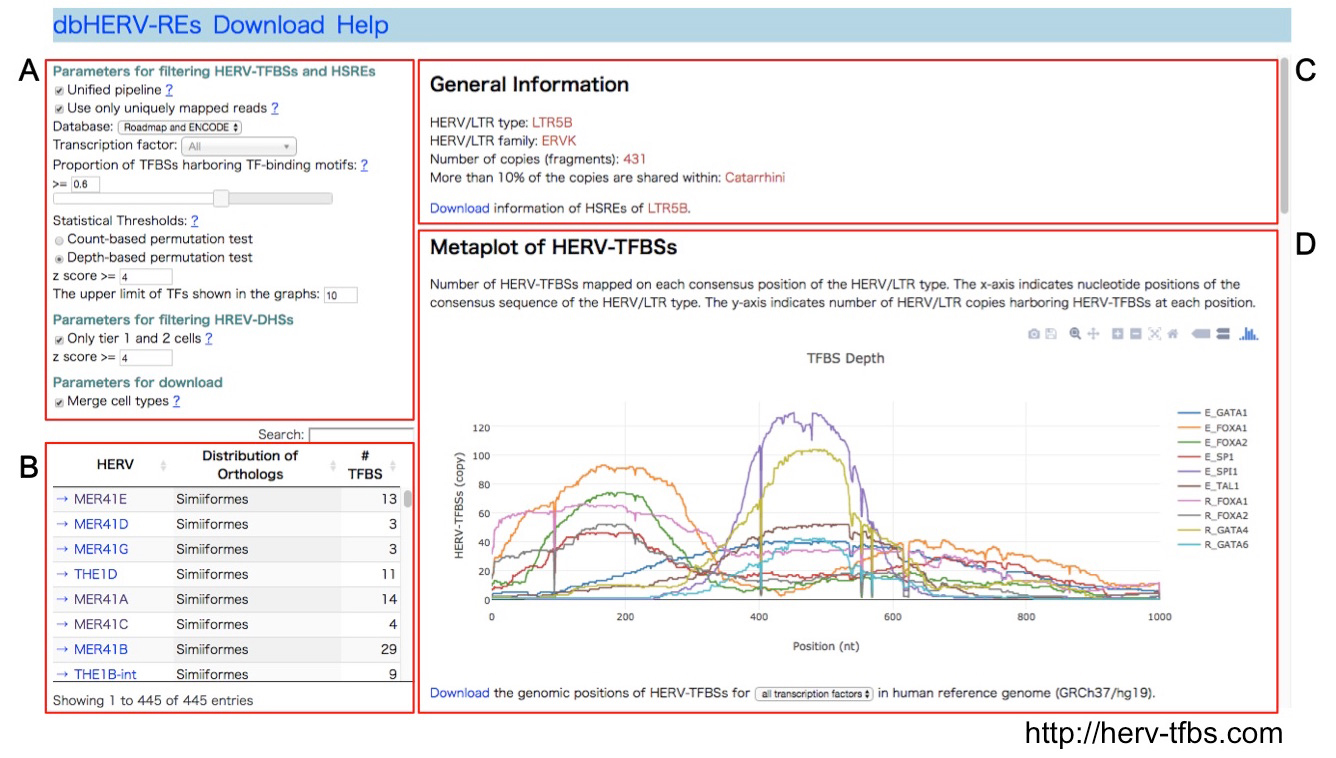
dbHERV-RE (http://herv-tfbs.com/). First, users choose a transcription factor and other parameters (A). Second, users select a type of HERVs (B). dbHERV-REs displays general information of the HERVs (phylogenetic classification, copy number, and insertion date) (C) and visualizes genetic positions of HERV-REs on the HERV sequence and the human reference genome (D).















