Dynamic nucleosome movement provides information on chromatin structure
![]()
Dynamic nucleosome movement provides structural information of topological chromatin domains in living human cells
Soya Shinkai, Tadasu Nozaki, Kazuhiro Maeshima, Yuichi Togashi
PLOS Computational Biology. 12(10), e1005136 DOI:10.1371/journal.pcbi.1005136
Pressrelease (In Japanese only)
The mammalian genome is organized into submegabase-sized chromatin domains (CDs) including topologically associating domains, which have been identified using chromosome conformation capture-based methods. Single-nucleosome imaging in living mammalian cells has revealed subdiffusively dynamic nucleosome movement (left in Fig.). It is unclear how single nucleosomes within CDs fluctuate and how the CD structure reflects the nucleosome movement. Here, we present a polymer model wherein CDs are characterized by fractal dimensions and the nucleosome fibers fluctuate in a viscoelastic medium with memory. We analytically show that the mean-squared displacement (MSD) of nucleosome fluctuations within CDs is subdiffusive. The diffusion coefficient and the subdiffusive exponent depend on the structural information of CDs. This analytical result enabled us to extract information from the single-nucleosome imaging data for HeLa cells (right in Fig.). Our observation that the MSD is lower at the nuclear periphery region than the interior region indicates that CDs in the heterochromatin-rich nuclear periphery region are more compact than those in the euchromatin-rich interior region with respect to the fractal dimensions as well as the size. Finally, we evaluated that the average size of CDs is in the range of 100–500 nm and that the relaxation time of nucleosome movement within CDs is a few seconds. Our results provide physical and dynamic insights into the genome architecture in living cells.
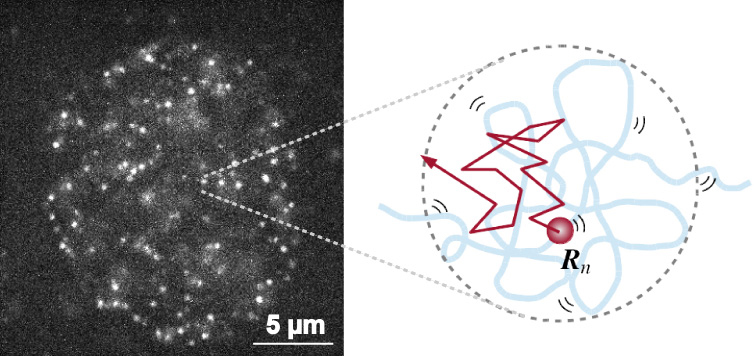
Single-nucleosome image in a living human nucleus (left). Nucleosome dynamically fluctuates within the chromatin domain and shows subdiffusion (right). By applying the mathematical model to the nucleosome movement, we can obtain the information on chromatin structure.















