Estimation of driving forces for cytoplasmic streaming using data assimilation
Cell Architecture Laboratory / Kimura Group
Bayesian Inference of Forces Causing Cytoplasmic Streaming in Caenorhabditis elegans Embryos and Mouse Oocytes.
Niwayama R., Nagao H., Kitajima T. S., Hufnagel L., Shinohara K., Higuchi T., Ishikawa T., Kimura A.
PLoS ONE, Vol 11, e0159917 (2016). DOI:10.1371/journal.pone.0159917
Cytoplasmic streaming is observed in wide variety of cells both in animals and plants. It is caused in many cases by cytoskeletons and molecular motors. Where these forces are exerted is difficult to identify. In this study, we developed a novel computational method to estimate the localization and amplitude of the forces generating cytoplasmic streaming. Our method infers the distribution of forces by fitting the flow field in hydrodynamics simulation to that observed in cells. We applied the method to Caenorhabditis elegans embryos and mouse oocytes. The distinct patterns of force distribution estimated in this study were consistent with the proposed distinct functions of the streaming in both of these species. We expect our method to have diverse applications, and to serve as a powerful tool for biologists who want to characterize the mechanics of biological hydrodynamic flows.
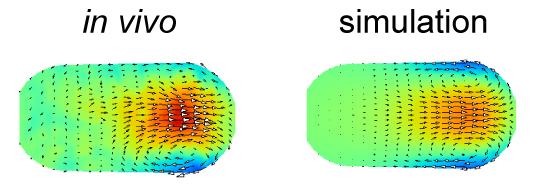
The velocity distribution in the simulation performed using the force distribution estimated in this study (right) agrees well with that measured experimentally (left). The color represents the velocity along the anterior-posterior axis, and the arrows represent the direction of the flow for cytoplasmic streaming in the C. elegans embryo.















