Emergence and evolution of Hominidae-specific coding and noncoding genomic sequences
Division of Population Genetics / Saitou Group
Emergence and evolution of Hominidae-specific coding and noncoding genomic sequences
Morteza Mahmoudi Saber, Isaac Adeyemi Babarinde, Nilmini Hettiarachchi and Naruya Saitou
Genome Biology and Evolution Volume 8, advance access, 2016 DOI:10.1093/gbe/evw132
Family Hominidae includes humans and great apes. We analyzed whole genome sequences to find Hominidae-specific genes and highly conserved noncoding sequences (HCNSs). We discovered that Down syndrome critical region 4 (DSCR4) is the only experimentally verified gene uniquely present in Hominidae. DSCR4 has no structural homology to any known protein and was inferred to have emerged in several steps. We also identified 1,658 Hominidae-specific HCNSs. These HCNSs were found to be under purifying selection, indicating that they may harbor important functions. They are in close proximity of genes involved in sensory perception of sound and developmental process, and also showed a significantly lower nucleosome occupancy probability. Interestingly, many ancestral sequences of the Hominidae-specific HCNSs showed very high evolutionary rates. This suggests that new functions emerged through some kind of positive selection, and then purifying selection started to operate to keep these functions.
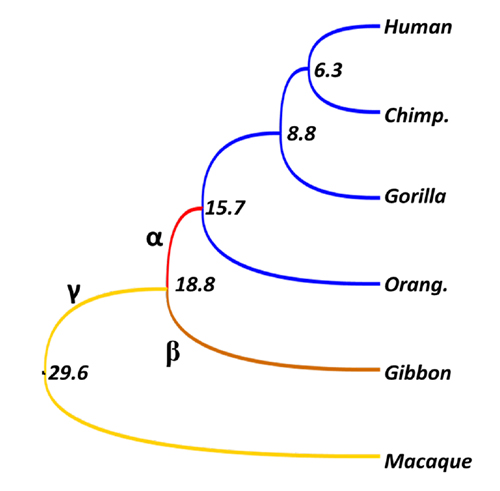
Evolutionary rates of each branch of Hominidae and their outgroups. Values are divergence times (million years). Although within-Hominidae rate is zero (identical sequences), rates for branches α, β, and γ 5.5, 2.0, and 1.9, respectively. In particular, the rate of branch α, which is common ancestor of Hominidae, is more than five times higher than neutral rate (1.0), and at least some ancestral sequences seem to experience positive selection.















