Genomic locations of conserved noncoding sequences and relationship with their function
Division of Population Genetics / Saitou Group
Genomic locations of conserved noncoding sequences and their proximal protein-coding genes in mammalian expression dynamics
Isaac Adeyemi Babarinde and Naruya Saitou
Molecular Biology and Evolution DOI:10.1093/molbev/msw058
Conserved Noncoding Sequences (CNSs) in mammals were examined with special reference to their genomic location. We extracted the CNSs conserved between chicken and four mammalian species (human, mouse, dog and cattle). Human CNSs were confirmed to be under purifying selection. The distribution pattern, ChIP-Seq and RNA-Seq data suggested that the CNSs are regulatory elements. Physical distances between CNS and their nearest protein coding genes were well conserved between human and mouse genomes. ChIP-Seq signal and gene expression patterns also suggested that CNSs regulate nearby genes. Genes with more CNSs have more evolutionarily conserved expression than those with fewer CNSs. These results suggest that the genomic locations of CNSs are important for their regulatory functions. In fact, various kinds of evolutionary constraints may be acting to maintain the genomic locations of CNSs and protein-coding genes in mammals to ensure proper regulation. First author of this paper, Dr. Babarinde, just receive Ph.D. from Department of Genetics, SOKENDAI. He also received Morishima Award from NIG.
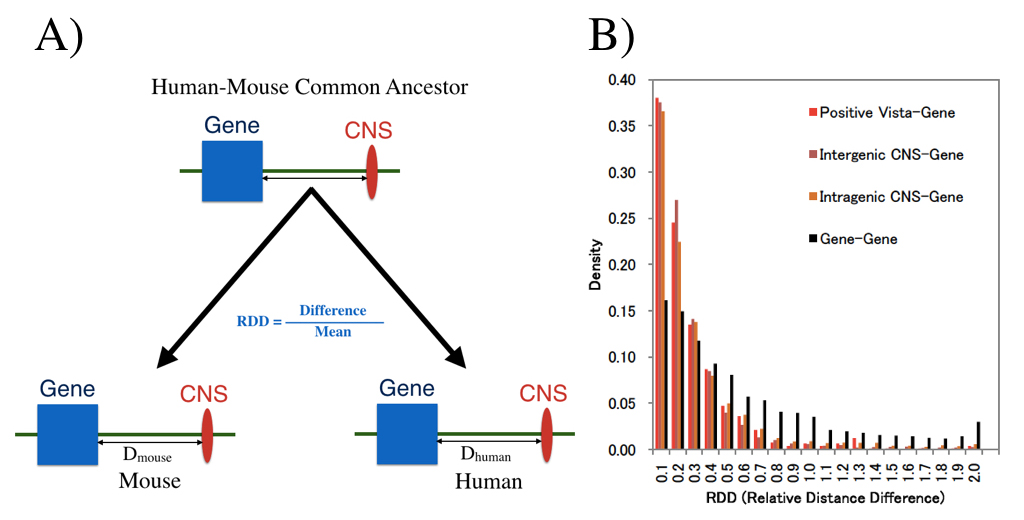
We examined whether genomic distances between genes and CNSs which already existed in the common ancestor of human and house are conserved now. (A) shows definition of distance measure RDD, and (B) shows RDD distribution. Distances between CNS and genes are much more conserved than those between genes.















