Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The RNA binding protein Nanos2 organizes a post-transcriptional buffering system to retain primitive states of mouse spermatogonial stem cells
Mammalian Development Laboratory / Saga Group
The RNA binding protein Nanos2 organizes a post-transcriptional buffering system to retain primitive states of mouse spermatogonial stem cells
Zhi Zhou, Takayuki Shirakawa, Kazuyuki Ohbo, Aiko Sada, Wu Quan, Kazuteru Hasegawa, Rie Saba, Yumiko Saga Developmental Cell. Published Online: June 25, 2015 DOI:http://dx.doi.org/10.1016/j.devcel.2015.05.014In many adult tissues, homeostasis relies on self-renewing stem cells that are primed for differentiation. The reconciliation mechanisms of these characteristics remain a fundamental question in stem cell biology. Here, we show that Nanos2, an evolutionarily conserved RNA-binding protein, works with other cellular messenger ribonucleoprotein (mRNP) components to ensure the primitive status of SSCs through a dual mechanism that involves 1) direct recruitment and translational repression of genes that promote spermatogonial differentiation, and 2) repression of the target of rapamycin complex 1 (mTORC1), a well-known negative pathway for SSC self-renewal, by sequestration of the core factor mTOR in mRNPs. This mechanism links mRNA turnover to mTORC1 signaling through Nanos2-containing mRNPs and establishes a post-transcriptional buffering system to facilitate SSC homeostasis in the fluctuating environment within the seminiferous tubule. This research is partly supported by Grant-in-Aid for Scientific Research on Innovative Areas ”Epigenome dynamics and regulation in germ cells” and a “Data assimilation” project of the Transdisciplinary Research Integration Center in ROIS. Zhi Zhou was a NIG postdoctoral fellow 2012.
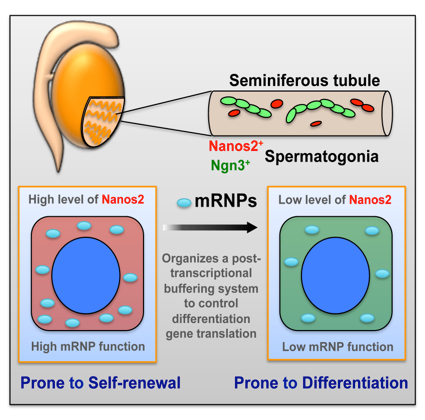
Nanos2 promotes mRNP formation and contribute to the maintenance of undifferentiated state of spermatogonial stem cells. Upon decreasing of Nanos2 level, stem cells shift to differentiation pathway.
Chemokine signalling controls integrity of radial glial scaffold in developing spinal cord and consequential proper position of boundary cap cells
Division of Brain Function / Hirata Group
Chemokine signalling controls integrity of radial glial scaffold in developing spinal cord and consequential proper position of boundary cap cells
Yan Zhu, Tomoko Matsumoto, Takashi Nagasawa, Fabienne Mackay, Fujio Murakami Journal of Neuroscience 17 June 2015, 35(24): 9211-9224; DOI:10.1523/JNEUROSCI.0156-15.2015Radial glial cells, the CNS neural progenitors, extend long radial processes that form the radial glial scaffold in the developing neuroepithelium. The integrity of this radial glial scaffold is well maintained throughout development. However, how this is achieved while the neuroepithelium rapidly expands and what the consequence might be when this integrity is compromised have not been well understood. In this study, we addressed these questions in the developing mouse spinal cord. We found that CXCR4, a receptor of a chemokine CXCL12 secreted from the pial meninges, is expressed in spinal cord radial glia. Conditional knockout of CXCR4 in radial glia causes disrupted radial glial scaffold with gaps at the pial endfeet layer, and consequentially lead to an invasion of boundary cap cells into the spinal cord. Since boundary cap cells are PNS cells normally positioned at the incoming and outgoing axonal roots, their invasion into the spinal cord suggests a compromised CNS/PNS boundary in the absence of CXCL12/CXCR4 signalling. By interrogating the underlying mechanisms, we then went on to show that CXCL12 signalling promotes the radial glia adhesion to basement membrane components and activates integrin β1 avidity. Our study uncovers an extrinsic molecular regulator for the maintenance of radial glial scaffold integrity during development. And our data suggest that the integrity of radial glia scaffold is important to safeguard the CNS/PNS boundary for the developing spinal cord.
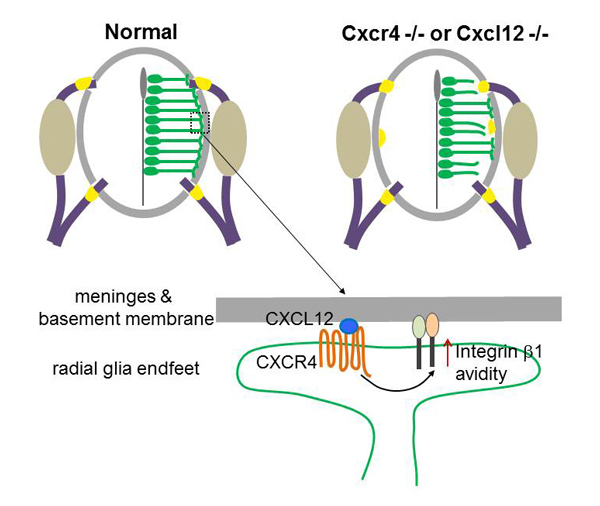
CXCL12 from the meninges regulates adhesion of radial glia to the basement membrane by enhancing avidity of integrin β1. Mice deficient in CXCL12 or its receptor CXCR4 show disrupted radial glial scaffold in their spinal cords, which consequentially causes invasion of boundary cap cells into the spinal cord.
User-friendly NIG mouse genome database
NIG_MoG (The National Institute of Genetics Mouse Genome database)
URL: http://molossinus.lab.nig.ac.jp/msmdb/
Mammalian Genetics Laboratory / Shiroishi Group
The National Institute of Genetics Mouse Genome database (NIG_MoG; http://molossinus.lab.nig.ac.jp/msmdb/) primarily comprises the whole genome sequence data of two inbred mouse strains, MSM/Ms and JF1/Ms. These strains were established at NIG and originated from the Japanese subspecies Mus musculus molossinus. A comparative analysis of their genomes with C57BL/6J (B6: whose genome is predominantly derived from the West European subspecies M. m. domesticus), revealed a vast number of SNPs, structural variants, insertions and deletions (indels). NIG_MoG provides, especially for wet-lab biologists, visualized genome polymorphism information, browsing single-nucleotide polymorphisms and short insertions and deletions in the genomes of MSM/Ms and JF1/Ms allowing users to intuitively recognize intersubspecific genome divergence in these mouse strains using visual data. The database also supports the in silico screening of BAC clones that contain genomic DNA from MSM/Ms and the standard classical laboratory strain C57BL/6N established at RIKEN BioResorce center. NIG_MoG is thus a valuable navigator for exploring mouse genome polymorphisms and BAC clones that are useful for studies of gene function and regulation based on intersubspecific genome divergence.
Fig. A screenshot of the top page of NIG_MoG
Sister DNA separation via compaction/expansion cycles
Biological Macromolecules Laboratory • Maeshima Group
Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles
Liang, Z., Zickler, D., Prentiss, M., Chang. F.S., Witz, G., Maeshima, K., Kleckner, N. Cell, 161, 1124-1137 (2015). DOI:10.1016/j.cell.2015.04.030Mammalian mitotic chromosome morphogenesis was analyzed by 4D live-cell and snapshot deconvolution fluorescence imaging. Prophase chromosomes, whose organization was previously unknown, are revealed to comprise co-oriented sister linear loop arrays displayed along a single, peripheral, regularly kinked topoisomeraseII /cohesin /condensinII axis (Fig a). Thereafter, rather than smooth, progressive compaction as generally envisioned, progression to metaphase is a discontinuous process involving chromosome expansion as well as compaction. At late prophase (Fig b), dependent on topoisomerase II and with concomitant cohesin release, chromosomes expand, axes split and straighten, and chromatin loops transit to a radial disposition around now-central axes. Finally, chromosomes globally compact, giving the metaphase state (Fig c). These patterns are consistent with the hypothesis that the molecular events of chromosome morphogenesis are governed by accumulation and release of chromosome stress, created by chromatin compaction and expansion. Chromosome state could evolve analogously throughout the cell cycle.
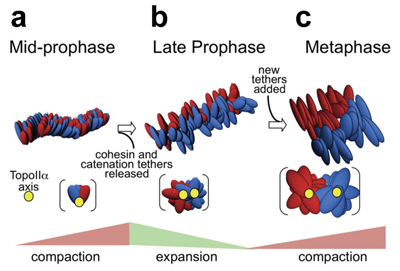
Two sister DNAs are shown in red and blue. The chromosome axis is in yellow. (a) mid-prophase; two sister chromatids are condensed with a single axis (yellow), but still mixed. (b) late-prophase; with expansion, two sisters and their axes are segregated. (c) metaphase; Further condensation and separation of the two sisters occur in metaphase.
OPEN HOUSE 2015
Time & Date
9:00 ~ 16:00, Saturday, April 4th (No Reservations Required, Free Admission)
Access
Free Shuttle Buses Available to the National Institute of Genetics from the North Exit of Mishima Station. ( Service Time: 8:50 ~ 15:00)
Map
Map Free Shuttle Buses Available Parking around Mishima Station Available for Car Visitors.
No Pets Allowed except Service Dogs, No General Parking*
(*Disabled Parking Available on the Institute Premises)
Information
1111 Yata, Mishima, Shizuoka 411-8540, JAPAN TEL:+81-55-981-5873















