Improvement of Pyrrole−Imidazole Polyamide Probes Targeting Human Telomeres
Biological Macromolecules Laboratory • Maeshima Group
(i) Structural Evaluation of Tandem Hairpin Pyrrole–Imidazole Polyam-ides Recognizing Human Telomeres
Hirata, A., Nokihara, K., Kawamoto, Y., Bando, T., Sasaki, A., Ide, S., Maeshima, K., Kasama, T., and Sugiyama, H. Journal of the American Chemical Society (JACS), July 18, 2014. DOI: 10.1021/ja506058e
(ii) Tandem Trimer Pyrrole-Imidazole Polyamide Probes Targeting 18 Base Pairs in Human Telomere Sequences
Kawamoto, Y., Sasaki, A., Hashiya, K., Ide, S., Bando, T. *, Maeshima, K. *, and Sugiyama, H.**co-corresponding authors
Chemical Science, January 20, 2015. DOI: 10.1039/C4SC03755C
Pyrrole-Imidazole (PI) polyamides bind to the minor groove of DNA in a sequence-specific manner. Our previous studies have demonstrated that a synthesized tandem hairpin PI polyamide (TH59) can target human telomere sequences (TTAGGG)n under mild conditions and can serve as a new probe for studying human telomeres. In the recently published two papers, we have improved specificity of the tandem hairpin PI polyamide by optimization of the connecting region (hinge region) of the tandem hairpin parts [HPTH59-b in Fig. A and paper (i)] and by introducing additional hairpin part [TT59 in Fig. B and paper (ii)]. The HPTH59-b and TT59 have higher specificity to recognize human telomeric repeats than the previous ones and stain telomeres in chemically fixed human cells with lower background signal.
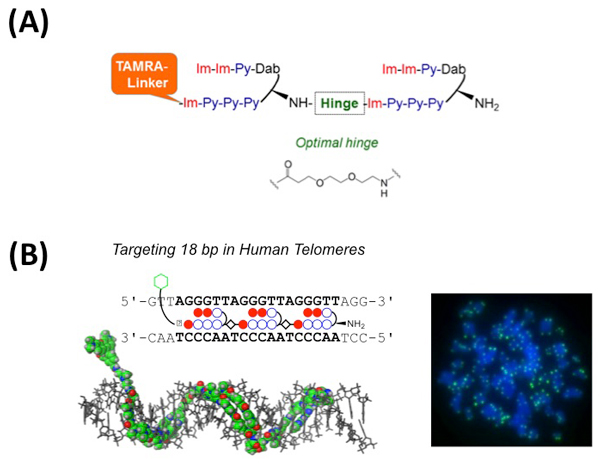
(A) Schematic structure of HPTH59-b. The optimized chemical structure of hinge segment is shown.
(B) Left, schematic representation of the tandem trimer PI polyamide TT59, which binds to 18bp of the human telomere sequence. Right, the telomere regions of chromosome ends are labeled with TT59 (green). DNA stain (blue).















