Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Muscle disease gene Stac3 identified in fish
Motor Neural Circuit Laboratory • Hirata Group
Horstick, E. J., Linsley, J. W., Dowling, J. J., Hauser, M. A., McDonald, K. K., Ashley-Koch, A., Saint-Amant, L., Satish, A., Cui, W. W., Zhou, W., Sprague, S. M., Stamm, D. S., Powell, C. M., Speer, M. C., Franzini-Armstrong, C., Hirata, H.* and Kuwada, J. Y.* (*Corresponding authors)
Nature Communications 4: 1952 (2013). doi:10.1038/ncomms2952
Native American myopathy (NAM) is a congenital muscular disease prevalent in the southern regions of the United States. However, the cause of this intractable myopathy has not been clarified so far. We originally studied mutant zebrafish that exhibited severe muscle weakness. The responsible gene encoded for a muscle protein Stac3. We found that Stac3 associates with dihydropyridine and ryanodine receptors and regulates calcium release during muscle contraction. Stac3-deficient zebrafish is now used for identifying drugs that mitigate the muscular defect in humans.
This is a collaborative work with Dr. John Y. Kuwada (University of Michigan).
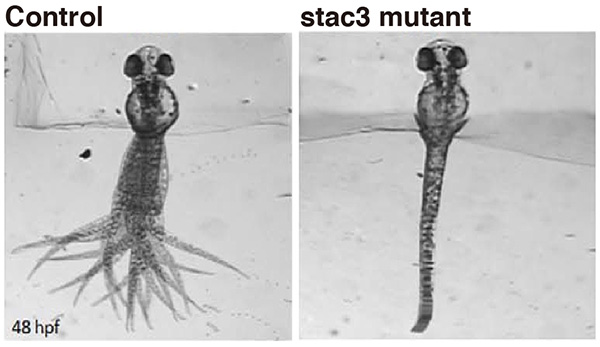
Touch evoked swimming in wild-type (control) but not stac3 mutant zebrafish embryos at 48 hours post-fertilization. Panels show superimposed frames of swimming motion with the head embedded in agarose.
ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability
Motor Neural Circuit Laboratory • Hirata Group
Hirata, H.*, Nanda, I.*, van Riesen, A.*, McMichael, G.*, Hu, H.*, Hambrock, M., Papon, M.-A., Fischer, U., Marouillat, S., Ding, C., Alirol, S., Bienek, M., Preisler-Adams, S., Grimme, A., Seelow, D., Webster, R., Haan, E., MacLennan, A., Stenzel, W., Yap, T. Y., Gardner, A., Nguyen, L. S., Shaw, M., Lebrun, N., Haas, S. A., Kress, W., Haaf, T., Schellenberger, E., Chelly, J., Viot, G., Shaffer, L. G., Rosenfeld, J. A., Kramer, N., Falk, R., El-Khechen, D., Escobar, L. F., Hennekam, R., Wieacker, P., Hübner, C., Ropers, H.-H., Gecz, J., Schuelke, M., Laumonnier, F. and Kalscheuer, V. M. (*Equal contribution)
American Journal of Human Genetics 92: 681-695 (2013). doi: 10.1016/j.bbr.2011.03.031
Arthrogryposis multiplex congenita (AMC) is caused by heterogeneous pathologies leading to multiple antenatal joint contractures through fetal akinesia. Understanding the pathophysiology of this disorder is important for clinical care of the affected individuals and genetic counseling of the families. In this study, we identified disease-causing mutations in the zinc-finger gene ZC4H2 in an AMC subtype that is associated with multiple dysmorphic features and intellectual disability (ID). In zebrafish, antisense-morpholino-mediated zc4h2 knockdown caused abnormal swimming and impaired primary motoneuron development. All missense mutations identified herein failed to rescue the swimming defect of zebrafish morphants. We conclude that ZC4H2 defects cause a clinically variable broad-spectrum neurodevelopmental disorder of the central and peripheral nervous systems. Our results highlight the importance of ZC4H2 for genetic testing of individuals presenting with ID plus muscle weakness and minor or major forms of AMC.
This is a collaborative work with Dr. Vera M. Kalscheuer (Max Planck Institute for Molecular Genetics), Dr. Markus Schuelke (Charité Universitätsmedizin Berlin) and other researchers and is supported by the NIG Collaborative Research.
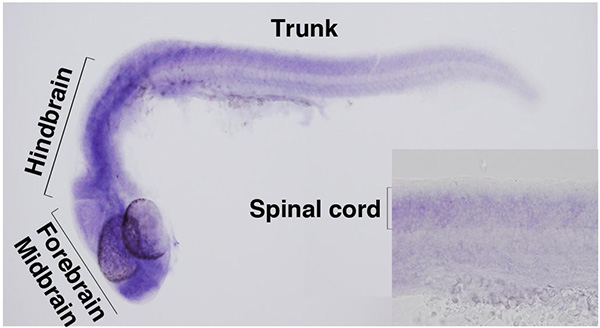
In zebrafish embryos, zc4h2 is expressed by forebrain, midbrain, hindbrain and spinal cord.
Mechanism for transcribing Arabidopsis genes containing intragenic heterochromatin
Division of Agricultural Genetics • Kakutani Group
Hidetoshi Saze, Junko Kitayama, Kazuya Takashima, Saori Miura, Yoshiko Harukawa, Tasuku Ito and Tetsuji Kakutani
Nature Communications, 4, Article number:2301 doi:10.1038/ncomms3301
Genomes of vertebrates and plants contain a substantial number of transposable elements (TEs), which are silenced by repressive epigenetic modifications, such as cytosine methylation and methylation of lysine 9 of histone H3. These modifications are essential for formation of inactive chromatin structures called heterochromatin. A potential complication is that active cellular genes sometimes contain TEs within their transcribed regions.
In this study, we show that heterochromatic epigenetic modifications are commonly found within actively transcribed gene units in both the Arabidopsis and rice genomes. We further show that in Arabidopsis, full-length transcription of genes with intragenic heterochromatin, most of which is formed by TE insertions, requires IBM2 (Increase in Bonsai Methylation 2), a protein with a Bromo-Adjacent Homology (BAH) domain and an RNA recognition motif (RRM). Our results reveal a novel epigenetic mechanism that masks effects of genetic variations created by TE insertions, allowing evolution of complex genomes with heterochromatic domains having diverse functions. (This work is a collaboration with Saze lab in OIST.)
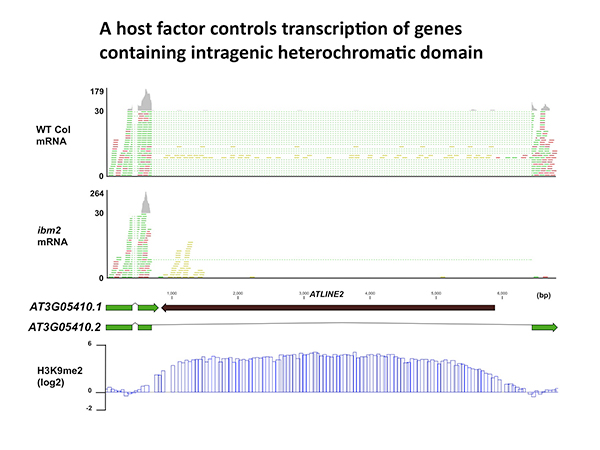
Full-length transcription of the gene containing heterochromatin is impaired in ibm2 mutant. mRNA reads are indicated with green and red. Reads with dotted lines are mapped across exon-exon boundaries. Note that the inserted TE (ATLINE2) is associated with heterochromatic H3K9 methylation (bottom).
Interhemispheric asymmetry of olfactory input-dependent neuronal specification in the adult zebrafish brain
Division of Molecular and Developmental Biology • Kawakami Group
Kishimoto, N., Asakawa, K., Madelaine, R., Blader, P., Kawakami, K., and Sawamoto, K.
Nature Neuroscience 16, 884-888, 2013 doi:10.1038/nn.3409
The vertebrate brain is anatomically and functionally asymmetric. The left and right cerebral hemispheres harbor neural stem cell niches at the ventricular-subventricular zone (V-SVZ) of the ventricular walls, where new neurons are continuously generated throughout life. However, any interhemispheric asymmetry of neural stem cell niches remains unclear. We performed gene-trap screens in adult zebrafish to identify genes that are differentially expressed in the two hemispheres and found that adult-born neurons expressing the neural zinc-finger protein Myt1 exist predominantly in the left V-SVZ. This lateralization could be reversed by left olfactory sensory deprivation–induced inactivation of Notch signaling. The olfactory behavioral preference for attractive amino acids was also impaired by sensory deprivation of the left olfactory system, but not of the right olfactory system. Our findings suggest that olfactory input generates interhemispheric differences in the fate of adult-born neurons in the zebrafish brain.
This study has been carried out as collaboration with Drs. Kishimoto and Sawamoto at Nagoya City University.
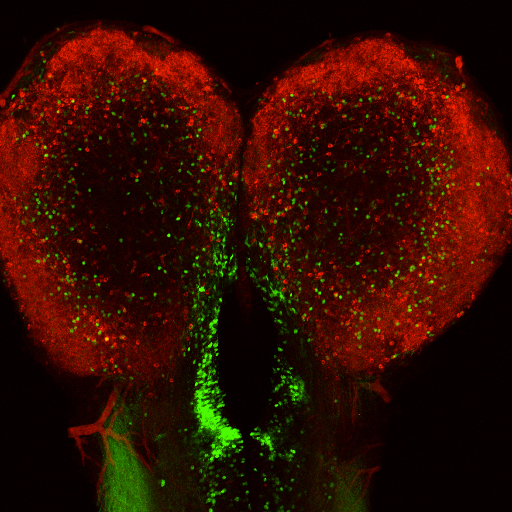
Interhemispheric asymmetric distribution of Myt1-positive neurons in telencephalic ventricular-subventricular zone.
Blood flow-dependent expression of miR-21 controls valvulogenesis of the zebrafish heart
Division of Molecular and Developmental Biology • Kawakami Group
Toshihiro Banjo,Janin Grajcarek,Daisuke Yoshino,Hideto Osada,Kota Y. Miyasaka,Yasuyuki S. Kida,Yosuke Ueki,Kazuaki Nagayama,Koichi Kawakami,Takeo Matsumoto,Masaaki Sato & Toshihiko Ogura
Nature Communications 4,Article number:1978,2013 doi:10.1038/ncomms2978
Heartbeat is required for normal development of the heart, and perturbation of intracardiac flow leads to morphological defects resembling congenital heart diseases. These observations implicate intracardiac haemodynamics in cardiogenesis, but the signalling cascades connecting physical forces, gene expression and morphogenesis are largely unknown. Here we use a zebrafish model to show that the microRNA, miR-21, is crucial for regulation of heart valve formation. Expression of miR-21 is rapidly switched on and off by blood flow. Vasoconstriction and increasing shear stress induce ectopic expression of miR-21 in the head vasculature and heart. Flow-dependent expression of mir-21 governs valvulogenesis by regulating the expression of the same targets as mouse/human miR-21 (sprouty, pdcd4, ptenb) and induces cell proliferation in the valve-forming endocardium at constrictions in the heart tube where shear stress is highest. We conclude that miR-21 is a central component of a flow-controlled mechanotransduction system in a physicogenetic regulatory loop.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
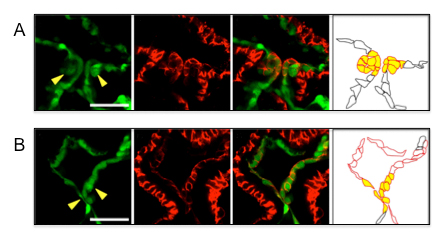
A: Normal valvulogenesis in the wild type zebrafish embryo.
B: Abnormal valvulogenesis in the zebrafish embryo in which the function of miR-21 was inhibited.
Recovery of rod-shaped cells by delayed cell division
Microbial Genetics Laboratory • Niki Group
Daisuke Shiomi, Hironori Niki
MicrobiologyOpen (DOI: 10.1002/mbo3.116)
RodZ is important for maintaining the rod shape of Escherichia coli. Loss of RodZ causes conversion of the rod shape to a round shape and a growth rate slower than that of wild-type cells. Suppressor mutations that simultaneously restore both the growth rates and the rod shape were isolated. Most of the suppressor mutations are found in mreB, mrdA, or mrdB. One of the mutations was in the promoter region of zipA, which encodes a crucial component of the cell division machinery. In this study, we investigated the mechanism of the suppression by this mutation. ZipA was slightly but significantly increased in the suppressor cells and led to a delay in cell division. While round-shaped mreB and mrdA mutants lose cell bipolarity, we found that round-shaped rodZ mutants retained cell bipolarity. Therefore, we concluded that a delay in the completion of septation provides extra time to elongate the cell laterally so that the zipA suppressor mutant is able to recover its ovoid or rod shape. The suppression by zipA demonstrates that the regulation of timing of septation potentially contributes to the conversion of morphology in bacterial cells.
This study has been carried out as collaboration with Dr. Ogura at Tohoku University.
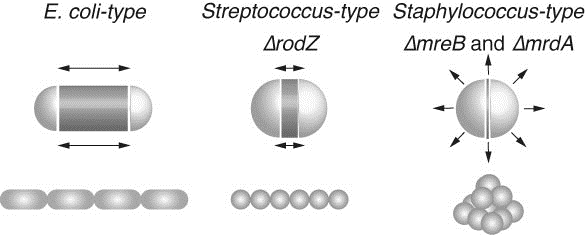
Rod shaped E. coli cells grow at the central cylinder. Ovoid shaped Streptococcus cells grow at septum while round shaped Staphylococcus cells swell. Black arrows indicate the direction of increase of cell volume. The increase in cell volume of ΔrodZ resembles that of Streptococcus, while those of ΔmreB or ΔmrdA resemble that of Staphylococcus. Dark gray zones indicate regions where peptidoglycan is actively synthesized. E. coli (WT), ΔrodZ, and Streptococcus cells retain cell polarity, while E. coli ΔmreB, ΔmrdA, and Staphylococcus cells lose polarity.
Chromosome condensation is affected by the size of the cell nucleus
Cell Architecture Laboratory • Kimura Group
Yuki Hara, Mari Iwabuchi, Keita Ohsumi, Akatsuki Kimura
Mol. Biol. Cell August 1, 2013 vol. 24(15) 2442-2453 doi: 10.1091/mbc.E13-01-0043
Chromosome condensation is critical for accurate inheritance of genetic information. The degree of condensation, which is reflected in the size of the condensed chromosomes during mitosis, is not constant. It is differentially regulated in embryonic and somatic cells. In addition to the developmentally programmed regulation of chromosome condensation, there may be adaptive regulation based on spatial parameters such as genomic length or cell size. We propose that chromosome condensation is affected by a spatial parameter called the chromosome amount per nuclear space, or “intranuclear DNA density.” Using Caenorhabditis elegans embryos, we show that condensed chromosome sizes vary during early embryogenesis. Of importance, changing DNA content to haploid or polyploid changes the condensed chromosome size, even at the same developmental stage. Condensed chromosome size correlates with interphase nuclear size. Finally, a reduction in nuclear size in a cell-free system from Xenopus laevis eggs resulted in reduced condensed chromosome sizes. These data support the hypothesis that intranuclear DNA density regulates chromosome condensation. This suggests an adaptive mode of chromosome condensation regulation in metazoans.
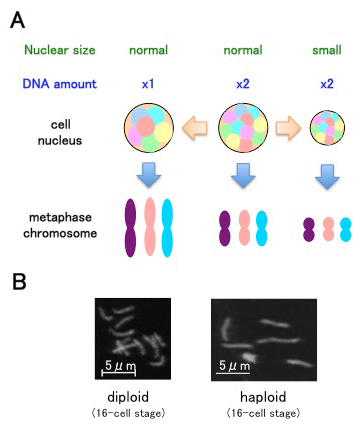
(A) Intranuclear DNA density affects chromosome condensation. When the nucleus is smaller, metaphase chromosomes are shorter. When the nucleus contains less chromosomal DNA, metaphase chromosomes are longer.
(B) Examples of metaphase chromosomes from C. elegans samples.















