Detailed analysis of a complex copy number variation region in mouse genome
Mouse Genomics Resource Laboratory (MGRL) • Koide Group
Umemori J, Mori A, Ichiyanagi K, Uno T, and Koide T.
BMC Genomics14: 455, 2013.
Copy number variation (CNV) of genomic segments is a common phenomenon that affects more than 10% of the human and mouse genomes, and is an important source of diversity in genomic structure. CNVs are frequently found in clusters called CNV regions (CNVRs), which are strongly associated with large segmental duplications (large SDs). However, the composition of these complex repetitive structures remains unclear. In the present study, we established new method for analyzing on complex repetitive structures of CNVRs by collaborating with National Institute of Informatics.
At first, we conducted self-comparative-plot analysis of all mouse chromosomes using the high-speed and large-scale-homology search algorithm, Similarity/Homology Efficient Analyze Procedure (SHEAP) developed by Dr. Takeaki Uno in National Institute of Informatics. By using this method, large SDs were visualized as unique tartan-checked patterns with complex arrangement of diagonal split lines (Figure 1). We focused on one SD on chromosome 13 (SD13M), which is one of the causative regions for genetic incompatibility (papers in preparation), and applied blast-based Systematic analysis of HErPlot to Extract Regional Distinction (SHEPHERD), a stepwise ab initio method, to extract core elements of repetitive sequences in SD13M (Figure 2). Then, comparative genome hybridization array analysis was empirically conducted on MSM, BLG2 (strains derived from wild mice in Mishima in Japan, and Toshevo in Bulgaria, respectively), and C57BL/6J (an experimental strain). This analysis showed that core elements are categorized to ones with CNVs and the others with constant copy number among strains, which have distinctive characters and divergences. The present study seems to be helpful for elucidating evolutional processes and functions of the CNVRs (Fig. 3).
This study was funded by the “Cultivation of integrated project” of Transdisciplinary Research Integration Center in Research Organization of Informatio and Systems (Japan).
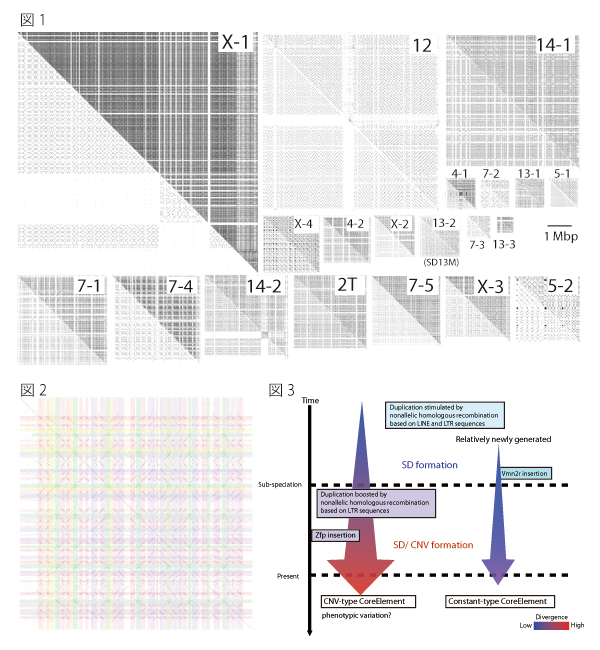
Fig.1. Tartan-checked structure of SDs visualized using SHEAP.
The lower left triangle of each panel shows a self-plot of the sequence after known repeat sequences have been masked using RepeatMasker. Each of the upper right triangles shows a self-plot of the intact sequence. Diagonal lines aligned in the same column or row represent repetitive sequences.
Fig.2. Extraction of repeat units from the self-plot of the large SD
Diagonal lines were extracted from a self-comparative-plot of SD13M that consisted of a dot-plot matrix. Then we selected one sequence and removed the other sequences represented by diagonal lines that were located in the same column or in the same row.
Fig.3 Model for the formation of SDs and CNVs
The average divergence of CNV-type core elements was greater than that of the constant type, and the CNV-type core elements contained significantly larger proportions of long terminal repeat (LTR) types of retrotransposon than the constant-type core elements. These results suggest that constant-type core elements emerged more recently than CNV-type core elements, and that retrotransposition of LTRs promotes nonallelic homologous recombination and caused CNV in SD13M.















