Okubo Group • Gene-Expression Analysis Laboratory
Developing technologies to make BioMedicine comprehensible and tractable
Faculty
Research Summary
A body of biomedical knowledge is developed in 2 steps: StepA: accumulating and exchanging situations captured in descriptions and data; and StepB: abstract situations into a coherent set of dogmatic or mathematical statements so that people can use in decision-making. The overwhelming output of A, mainly due to the technological assistance by diagnostic, laboratory and communication machines, is making a stressful situation called “information over-load” or “data deluge”. New technologies must be invented for B to make bigger return from investment in biomedicine.
Selected Publications
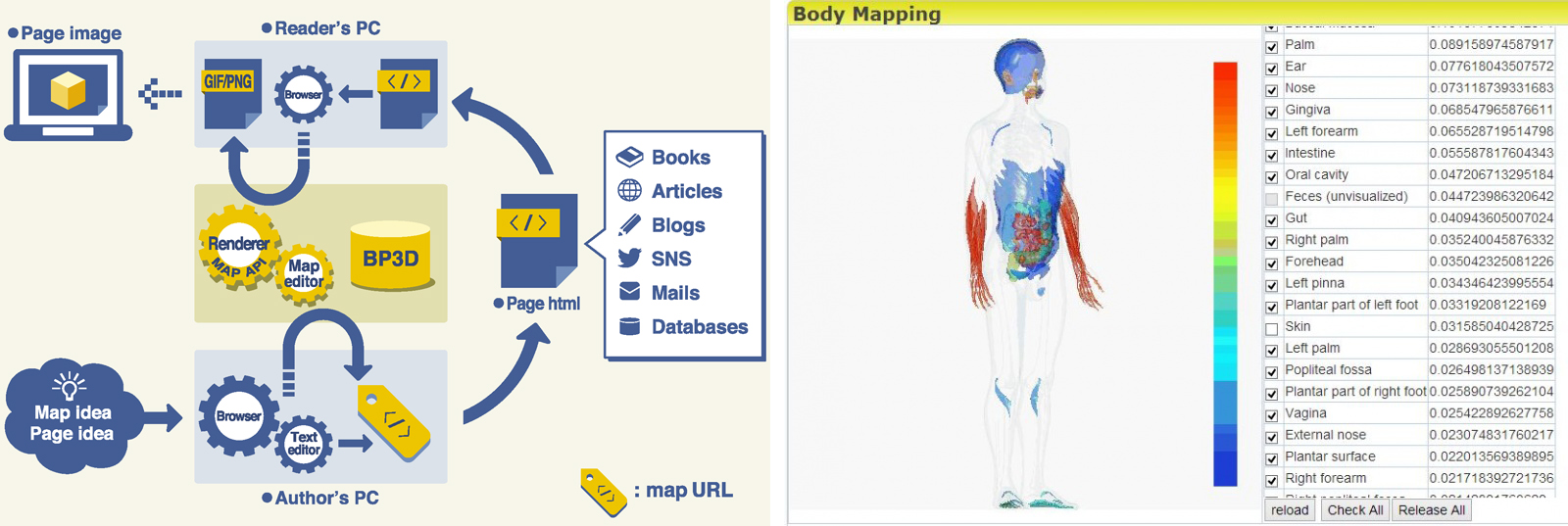
Fujieda K, Okubo K. A reusable anatomically segmented digital mannequin for public health communication. J Vis Commun Med. 2016 Jan-Jun;39(1-2):18-26.
Ono H, Ogasawara O, Okubo K, Bono H. RefEx, a reference gene expression dataset as a web tool for the functional analysis of genes. Sci Data. 2017 Aug 29;4:170105.
Mitsuhashi N, Fujieda K, Tamura T, Kawamoto S, Takagi T, Okubo K. BodyParts3D: 3D structure database for anatomical concepts. Nucleic Acids Res. 2009 Jan;37(Database issue):D782-5.
Okubo K, Tamura T. System and computer software program for visibly processing an observed information’s relationship with knowledge accumulations. 2011 US patent 20050203889
















