A small RNA system is involved in homology recognition between meiotic chromosomes
Press release
A small RNA system ensures accurate homologous pairing and unpaired silencing of meiotic chromosomes
Hiroaki Tabara*, Shohei Mitani, Megumi Mochizuki, Yuji Kohara and Kyosuke Nagata *: corresponding author
The EMBO Journal 2023 April 20 DOI:10.15252/embj.2020105002
Meiosis is a specialized mode of cell division that produces haploid gametes. During meiosis, chromosomes with homologous partners undergo synaptonemal complex (SC)-mediated pairing, while the remaining unpaired regions such as most parts of the sex chromosomes in males are heterochromatinized through unpaired silencing.
A study, published in The EMBO Journal, introduces that a small RNA system regulates meiotic pairing of homologous chromosomes in the nematode Caenorhabditis elegans. Homolog pairing initiation depends on species-specific mechanisms involving interactions between pericentric heterochromatin, centromeres, telomeres, and/or pairing center sequences. Subsequently, the SC establishes the pairing of other regions of the homologous chromosomes. In plants and mammals, DNA double-strand breaks (DSBs) are important or partly important for synapse formation between homologous chromosomes. Mysteriously, in C. elegans and fruit fly, DSBs are not necessary for synapse formation, implying the presence of an unclarified mechanism for recognizing homologous chromosomes during SC formation.
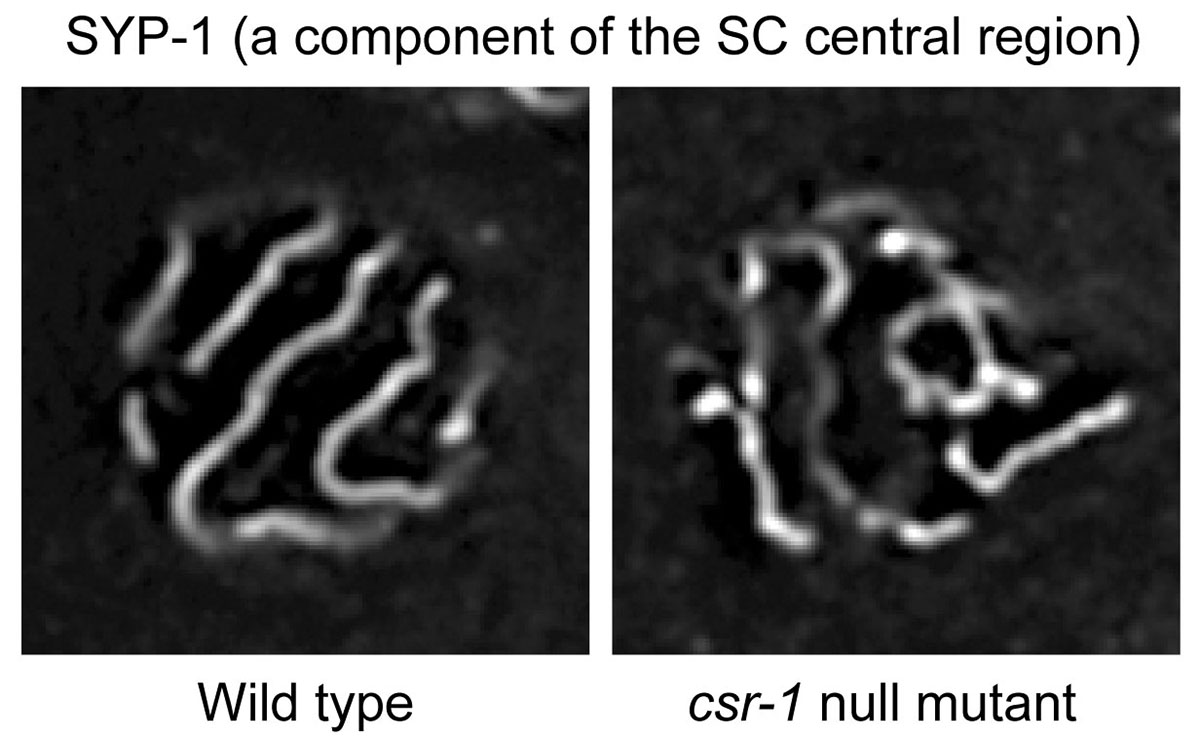
RNA interference (RNAi) is a form of RNA-mediated sequence-specific regulation of gene expression in eukaryotes. Small RNAs and Argonaute proteins play key roles in the mechanism of RNAi. Researchers at National Institute of Genetics, Tokyo Women’s Medical University, and University of Tsukuba, in Japan, found new relationships between SC formation on paired chromosomes, unpaired chromosome heterochromatinization, and an Argonaute–small RNA pathway (endogenous RNAi). The C. elegans Argonaute proteins, CSR-1 and its paralog interacting with small RNAs, are required for SC formation between chromosome pairs with accurate homology and are also involved in the condensation of unpaired chromosomes. The SC is composed of layers of strings, with two lateral elements sandwiching a central region. CSR-1 in nuclei and meiotic cohesin, constituting the SC lateral elements, were associated with non-simple DNA repeats expressing small RNAs and long non-coding RNAs (lncRNAs), and also associated weakly with coding genes. This study proposes a zipper model, in which the accurate homolog pairing is promoted via the inter-chromosomal recognition of chromosome-associated lncRNAs by the Argonaute-mediated small RNA system. The meiotic difference between paired or unpaired chromosomes is also described.
Reference
Tabara H, Mitani S, Mochizuki M, Kohara Y, Nagata K (2023) A small RNA system ensures accurate homologous pairing and unpaired silencing of meiotic chromosomes. EMBO J, in press. DOI: 10.15252/embj.2020105002.