Functional diversity of gibberellin activation enzymes leading to interspecific differences in pollen tube germination/elongation among Oryza species
Evolutionary alterations in gene expression and enzymatic activities of gibberellin 3-oxidase 1 in Oryza
Kyosuke Kawai, Sayaka Takehara, Toru Kashio, Minami Morii, Akihiko Sugihara, Hisako Yoshimura, Aya Ito, Masako Hattori, Yosuke Toda, Mikiko Kojima, Yumiko Takebayashi, Hiroyasu Furuumi, Ken-ichi Nonomura, Bunzo Mikami, Takashi Akagi, Hitoshi Sakakibara, Hidemi Kitano, Makoto Matsuoka & Miyako Ueguchi-Tanaka
Communications Biology (2022) 5, 67 DOI:10.1038/s42003-022-03008-5
The plant hormone gibberellin (GA) plays important roles in various developmental events, such as stem elongation, induction of seed germination and flowering. Although GA is indispensable for anther and pollen development, our knowledge of GA functions during plant reproduction has been limited to date.
This paper reports the functional and evolutionary analyses of rice gibberellin 3-oxidase 1 (OsGA3ox1), a gibberellin synthetic enzyme specifically expressed in the late developmental stages of anthers. Enzymatic and X-ray crystallography analyses reveal that OsGA3ox1 has a higher GA7 synthesis ratio than OsGA3ox2. In addition, we generate an osga3ox1 knockout mutant by genome editing and demonstrate the bioactive gibberellic acid synthesis by the OsGA3ox1 action during starch accumulation in pollen via invertase regulation. Furthermore, we analyze the evolution of Oryza GA3ox1s and reveal that their enzyme activity and gene expression have evolved in a way that is characteristic of the Oryza genus and contribute to their male reproduction ability.
In this paper, we used the wild strains of genus Oryza that NIG has conserved with the support of National Bioresource Project (NBRP) Rice, Ministry of Education, Culture, Sports, Science and Texhnology (MEXT), Japan.
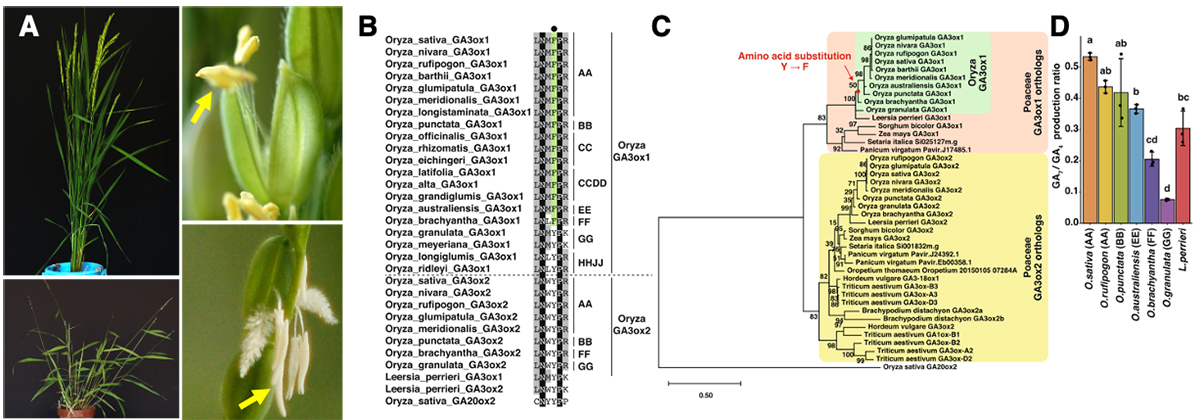
(A) Plant types and florets of cultivated species, Oryza sativa (top), and wild relatives, O. granulata (bottom). Yellow arrows indicates anthers. (B) Amino acid sequence alignment in the 2OG-interacting site around F (in OsGA3ox1) of Poaceae GA3ox orthologs and OsGA20ox2. Residues of F in the interacting site of 2OG are indicated in green, those that are 100% identical are shown in black, and those with more than 50% identity are depicted in gray. The black dot indicates the amino acid in the interacting site with co-substrate 2OG, F (shown in green), or Y. On the right side of the alignment, the letters AA and BB indicate the AA and BB Oryza genomes, respectively. (C) Phylogenetic tree of Poaceae GA3ox orthologs and OsGA20ox2. The red dot indicates the common ancestor of O. sativa to O. brachyantha that acquired the residue F in the active site. (D) In vitro enzyme activity of GA3ox1 of Oryza species and L. perrieri; O. sativa GA3ox1, O. rufipogon GA3ox1, O. punctata GA3ox1, O. brachyantha GA3ox1, O. granulata GA3ox1, L. perrieri GA3ox1. Error bars, s.d. n = 3, one-way ANOVA with Tukey’s multiple comparisons test. Different letters denote significant differences (p < 0.05).















