Telomere-specific chromatin capture using a pyrrole–imidazole polyamide probe
Maeshima Group / Genome Dynamics Laboratory
Telomere-specific chromatin capture using a pyrrole–imidazole polyamide probe for the identification of proteins and non-coding RNAs
Satoru Ide#*, Asuka Sasaki#, Yusuke Kawamoto, Toshikazu Bando, Hiroshi Sugiyama, Kazuhiro Maeshima
#Equally contributed, *Corresponding author
Epigenetics & Chromatin (2021) 14, 46 DOI:10.1186/s13072-021-00421-8
Background: Knowing chromatin components at a DNA regulatory element at any given time is essential for understanding how the element works during cellular proliferation, differentiation and development. A region-specific chromatin purification is an invaluable approach to dissecting the comprehensive chromatin composition at a particular region. Several methods (e.g., PICh, enChIP, CAPTURE and CLASP) have been developed for isolating and analyzing chromatin components. However, all of them have some shortcomings in identifying non-coding RNA associated with DNA regulatory elements.
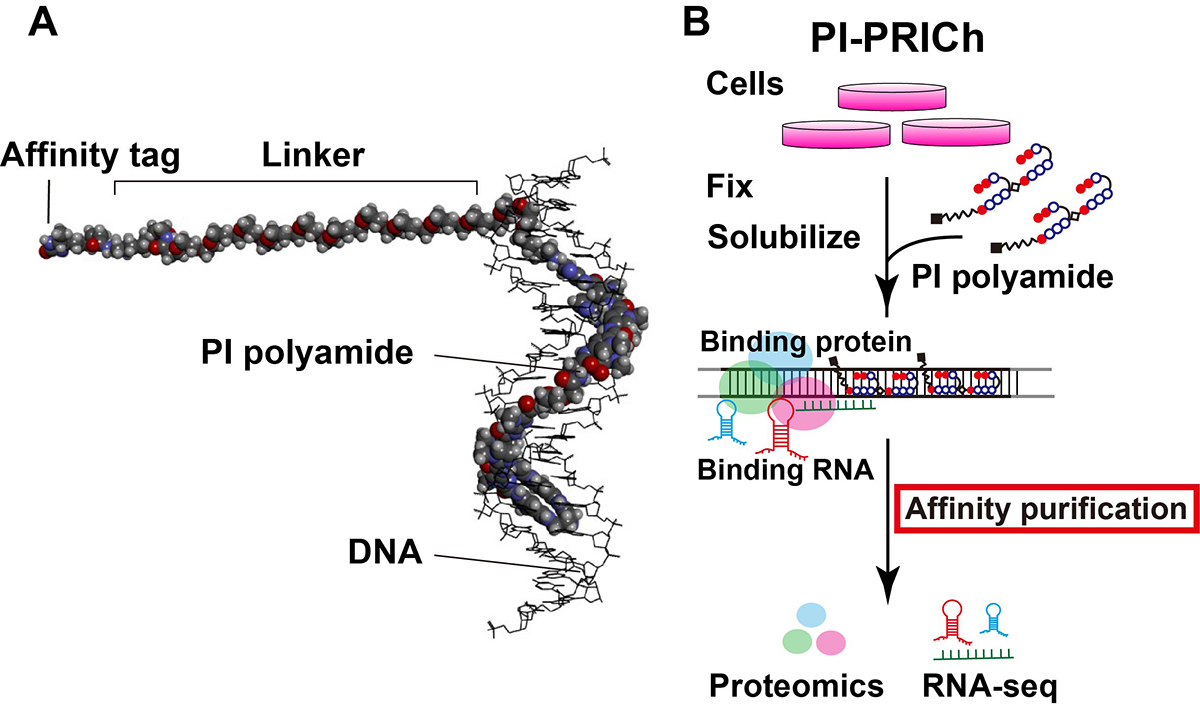
Results: We have developed a new approach for affinity purification of specific chromatin segments employing an N-methyl pyrrole (P)−N-methylimidazole (I) (PI) polyamide probe, which binds to a specific sequence in double-stranded DNA via Watson–Crick base pairing as a minor groove binder (Figure 1A). This new technique is called proteomics and RNA-omics of isolated chromatin segments (PI-PRICh). Using PI-PRICh to isolate mouse and human telomeric components, we found enrichments of shelterin proteins, the well-known telomerase RNA component (TERC) and telomeric repeat-containing RNA (TERRA) When PI-PRICh was performed for alternative lengthening of telomere (ALT) cells with highly recombinogenic telomeres, in addition to the conventional telomeric chromatin, we obtained chromatin regions containing telomeric repeat insertions scattered in the genome and their associated RNAs.
Conclusion: PI-PRICh reproducibly identified both the protein and RNA components of telomeric chromatin when targeting telomere repeats. PI polyamide is a promising alternative to simultaneously isolate associated proteins and RNAs of sequence-specific chromatin regions under native conditions, allowing better understanding of chromatin organization and functions within the cell (Figure 1B).
This work was supported by an NIG-JOINT (2015-B6), JSPS grants (JP17J10836 to A.S.; 15H01361 and 21H02535 to S.I.; 20H05936 and 21H02453 to K.M.), the Takeda Science Foundation to K.M. and the Uehara Memorial Foundation to K.M.. A.S. was a JSPS Fellow (DC2).