Condensin II makes the chromosome territory in the nucleus
3D genomics across the tree of life reveals condensin II as a determinant of architecture type
Hoencamp, C., Dudchenko, O., Elbatsh, A. M.O., Brahmachari, S., Raaijmakers, J. A., van Schaik, T., Cacciatore, Á. S., Contessoto, V., van Heesbeen, R. G.H.P., van den Broek, B., Mhaskar, A. N., Teunissen, H., St Hilaire, B. G., Weisz, D., Omer, A. D., Pham, M., Colaric, Z., Yang, Z., Rao, S. S.P., Mitra, N., Lui, C., Yao, W., Khan, R., Moroz, L. L., Kohn, A., St. Leger, J., Mena, A., Holcroft, K., Gambetta, M. C., Lim, F., Farley, E., Stein, N., Haddad, A., Chauss, D., Mutlu, A. S., Wang, M. C., Young, N. D., Hildebrandt, E., Cheng, H. H., Knight, C. J., Burnham, T. L.U., Hovel, K. A., Beel, A. J., Mattei, P.-J., Kornberg, R. D., Warren, W. C., Cary, G., Gómez-Skarmeta, J. L., Hinman, V., Lindblad-Toh, K., di Palma, F., Maeshima, K., Multani, A. S., Pathak, S., Nel-Themaat, L., Behringer, R. R., Kaur, P., Medema, R. H., van Steensel, B., de Wit, E., Onuchic, J. N., Di Pierro, M., Lieberman-Aiden, E., Rowland, B. D.
Science 372, 984-989 (2021) DOI:10.1126/science.abe2218
An international collaborative team, which was led by B.D. Rowland at the Netherlands Cancer Institute and E. Lieberman Aiden (NIG International Strategic Advisor) at Baylor College of Medicine, investigated genome folding across the eukaryotic tree of life (24 species, Fig. B). The team found four types of 3D genome architecture at chromosome-scale (Fig. A). Each type appeared and disappeared repeatedly during eukaryotic evolution. The type of genome architecture that an organism exhibits correlates with the absence of condensin II subunits (Fig. B). Condensin is a protein complex required for mitotic chromosome assembly. In vertebrates, it is known that there are two types of condensins, condensin I and II, and that condensin II shortens mitotic chromosomes.
Moreover, the team demonstrated that condensin II depletion converted the architecture of the human genome to a state resembling that seen in organisms such as fungi or mosquitoes (Rabl-like, Fig. C). In this state, centromeres clustered together at nucleoli, and heterochromatin domains merged. The team proposed a physical model in which lengthwise compaction of chromosomes by condensin II during mitosis determines chromosome-scale genome architecture, with effects that are retained during the subsequent interphase. This mechanism is likely conserved since the last common ancestor of all eukaryotes.
The part to which NIG contributed was supported by MEXT KAKENHI grants (20H05936).
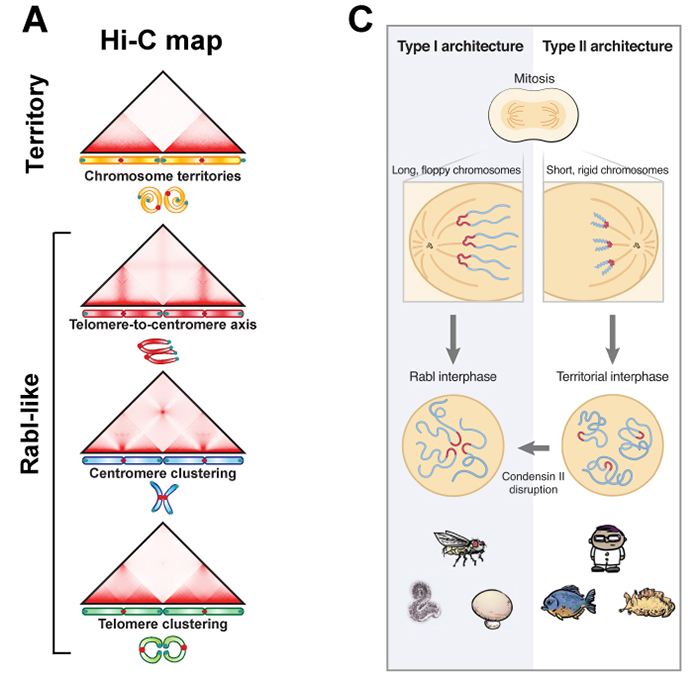
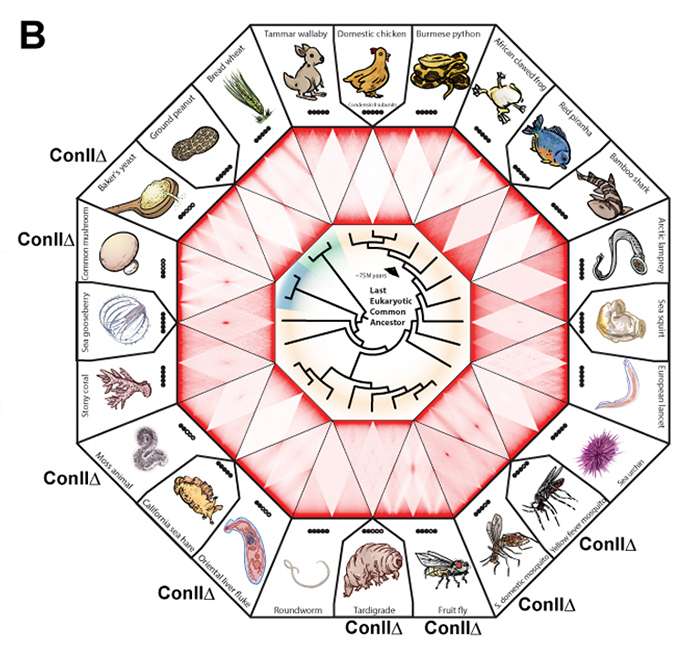
Figure: (A) Chromosome positioning in the nucleus classified by Hi-C maps: “chromosome territory” and “rabl-like”. (B) Hi-C maps of 24 species covering animals (yellow), fungi (blue) and plants (green). Presence of the condensin II subunits in each species is indicated by solid black circles. In case of condensin II-deficiency, shown as “Con IIΔ”. (C) Model. Having shorter chromosomes during cell division leads to separate centromeres and territorial genome architecture in the subsequent interphase. Reducing chromosome lengthwise compaction by condensin II disruption, leads to enhanced centromere clustering, loss of chromosome territories, and a Rabl-like genome architecture.















