Mechanisms to stabilize genome by chromatin remodeling
Kakutani Group / Epigenomics Laboratory
The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W
Akihisa Osakabe, Bhagyshree Jamge, Elin Axelsson, Sean A. Montgomery, Svetlana Akimcheva, Annika Luisa Kuehn, Rahul Pisupati, Zdravko J. Lorković, Ramesh Yelagandula, Tetsuji Kakutani, and Frédéric Berger
Nature Cell Biology 23, 391-400 (2021) DOI:10.1038/s41556-021-00658-1
Transposable elements (TEs) are generally silenced by epigenetic modifications such as methylation of DNA and histone. An Arabidopsis chromatin remodeler protein DDM1 (Decrease in DNA methylation 1) was identified by genetic screening as a factor necessary for maintenance of these silencing marks. However, the underlying mechanisms of this function of DDM1 remain unknown.
In this study, we focused on histone variant, H2A.W, which localizes at TE-rich heterochromatic regions. Our genomic analysis revealed that H2A.W was lost from heterochromatin when DDM1 is non-functional. DDM1 directly binds to H2A.W. Histone binding regions of DDM1 are important for H2A.W deposition and TE silencing. These results suggest that DDM1 silences TEs by deposition of H2A.W into heterochromatin. This pathway to silence TEs by chromatin remodeling could be conserved to mammals, since mammals have DDM1 and H2A.W orthologs with similar functions.
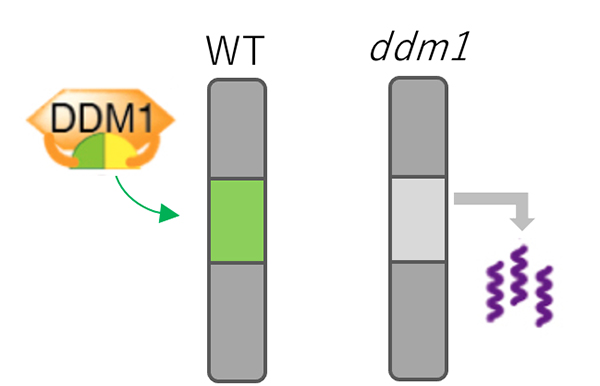
Figure: Mechanism for TE silencing regulated by DDM1.
In wild type plant (left), DDM1 mediates deposition of H2A.W (shown by green) into genomic regions with silent TEs. In ddm1 mutants (right), H2A.W was lost from heterochromatin, which is associated with derepression of TEs.















