Mechanisms to silence mobile DNA within the genome
RNA interference-independent reprogramming of DNA methylation in Arabidopsis
Taiko Kim To, Yuichiro Nishizawa, Soichi Inagaki, Yoshiaki Tarutani, Sayaka Tominaga, Atsushi Toyoda, Asao Fujiyama, Frédéric Berger, Tetsuji Kakutani
Nature Plants 2020 November 30 DOI:10.1038/s41477-020-00810-z
DNA methylation is important for silencing transposable elements (TEs) in diverse eukaryotes including plants. In plant genomes, TEs are silenced by methylation of histone H3 lysine 9 (H3K9) and cytosines in both CG and non-CG contexts. The role of RNA interference (RNAi) in establishing TE-specific silent marks has been extensively studied, but the significance of RNAi-independent pathways remains largely unexplored. Here, we directly investigated transgenerational de novo DNA methylation of TEs after the loss of silent marks. Our analyses uncovered potent and precise RNAi-independent pathways for recovering non-CG methylation and H3K9 methylation in most TE genes (i.e. coding regions within TEs). Characterization of a subset of TE genes without the recovery revealed the impacts of H3K9 demethylation, replacement of histone H2A variants, and their interaction with CG methylation, together with feedback from transcription. These chromatin components are conserved among eukaryotes and may contribute to chromatin reprograming in a conserved manner.
Supported by Systems Functional Genetics Project of the Transdisciplinary Research Integration Center, ROIS.
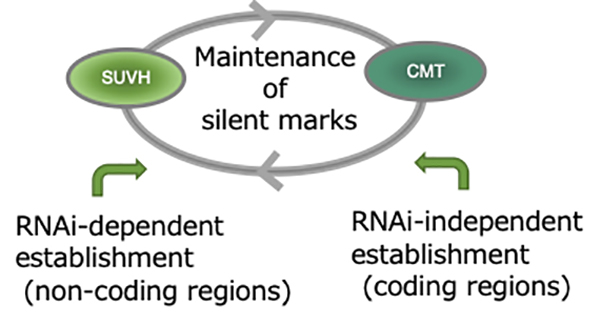
Figure: H3K9 and non-CG methylation can be maintained and inherited by mutual reinforcement. These marks are established by RNAi-dependent and -independent pathways. The RNAi-independent pathway mainly works in coding regions of TEs.















