Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Homecoming Visit and Gatherings During the Year-End and New Year Holidays
There are less than ten days left this year. Some people are likely to return home or hold gatherings during the year-end and New Year holidays.
Unfortunately, the third wave of the novel coronavirus pandemic has not yet been tamed. It has even grown increasingly severe. If we return to our hometown at a time like this, the infection risk will increase as there will be occasions to meet or to have meals together with elderly family members and with friends whom we seldom meet. If we join a year-end party or a New Year party, we are inclined to spend quite a long time speaking without a mask while dining, and will have a greatly increased risk of infection.
We are at a crucial time now. NIG members will refrain from returning to hometowns during the year-end and New Year Holidays as much as possible. If it is unavoidably necessary to visit home, we will make sure to wear a mask, disinfect hands, avoid speaking in a loud voice, avoid three conditions of “poor ventilation” “intensive crowds” and “close social distance”, and ventilate closed spaces while on the way to and from home and during the stay. When visiting public facilities, we will take utmost precautions against infection and wear a mask out of doors, too. We will also monitor our daily health condition using the health observation form.
We are going to take a break on the holidays, but there is no break for the novel coronavirus!!
We will consider carefully whether it is worth the risk to go ahead with homecoming/gathering plans and act cautiously wherever possible. Let’s greet the coming year healthily and soundly.
Best regards,
Fumio Hanaoka
Director-General (Head of the Department of Genetics)
National Institute of Genetics
December 23, 2020
Changes in gene expression in the brain are involved in domestication of mice
Combined Change of Behavioral Traits for Domestication and Gene‐Networks in Mice Selectively Bred for Active Tameness
Yuki Matsumoto, Hiromichi Nagayama, Hirofumi Nakaoka, Atsushi Toyoda, Tatsuhiko Goto, Tsuyoshi Koide
Genes Brain Behav. 2020 December 13 DOI:10.1111/gbb.12721
Tameness can be divided into two components: active tameness and passive tameness. We conducted selective breeding for higher scores of active tameness using a wild-derived heterogeneous stock. In the analyses of nine behavioral traits related to tameness, we found that five traits showed changes in the selective groups compared to the control groups through the generations. We conducted cluster analyses to evaluate the relationship among the nine traits and RNA-Seq analysis to characterize the molecular network related to tameness. The results suggested that active tameness was hidden in the control groups but became apparent in the selected populations by selective breeding, potentially driven by changes in gene expression networks.
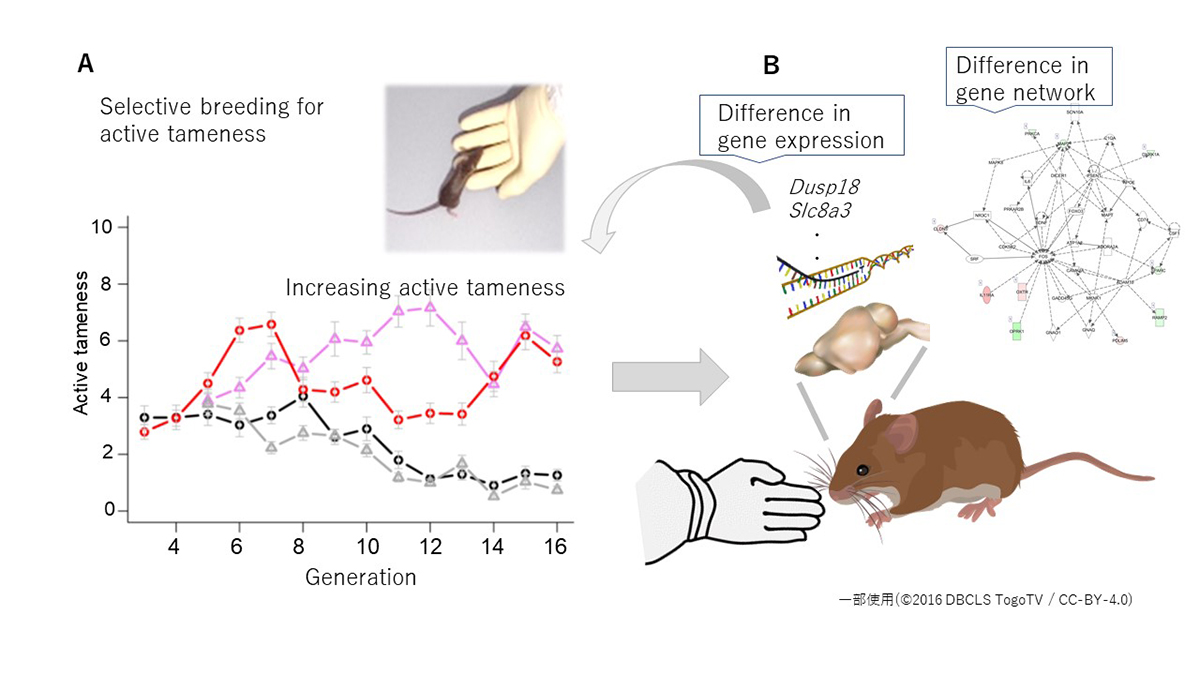
Figure: The wild-derived heterogeneous stock was divided into four groups, two groups were selected for active tameness, and the other two groups were unselected controls. A: As the generation progressed, active tameness increased in the two selection groups, whereas there was no significant increase in the two control groups. B: RNA-seq analysis was performed using samples collected from the brains (hippocampus) of mice in the selection groups and the control groups. As a result, genes with different expression levels were found in the selection groups and the control groups, and differences in the gene network were also found.
A protein maintaining plastid and mitochondrial genome integrity in plants
Holliday junction resolvase MOC1 maintains plastid and mitochondrial genome integrity in algae and bryophytes
Yusuke Kobayashi, Masaki Odahara, Yasuhiko Sekine, Takashi Hamaji, Sumire Fujiwara, Yoshiki Nishimura, Shin-ya Miyagishima
Plant Physiology 184, 1870-1883 (2020) DOI:10.1104/pp.20.00763
When DNA double-strand breaks occur, four-stranded DNA structures called Holliday junctions (HJs) form during homologous recombination (HR). Because HJs connect homologous DNA by a covalent link, resolution of HJ is crucial to terminate HR and segregate the pair of DNA molecules faithfully. We recently identified Monokaryotic Chloroplast 1 (MOC1) as a plastid DNA (ptDNA) HJ resolvase in algae and plants. Although Cruciform cutting endonuclease 1 (CCE1) was identified
as a mitochondrial DNA (mtDNA) HJ resolvase in yeasts, homologs or other mitochondrial HJ resolvases have not been identified in other eukaryotes. Here, we demonstrated that MOC1 depletion in the green alga Chlamydomonas reinhardtii and the moss Physcomitrella patens induced ectopic recombination between short dispersed repeats (SDRs) in ptDNA. In addition, MOC1 depletion disorganized thylakoid membranes in plastids. In some land plant lineages, such as the moss P. patens, a liverwort and a fern, MOC1 dually targeted to plastids and mitochondria. Moreover, mitochondrial targeting of MOC1 was also predicted in charophyte algae and some land plant species. Besides causing instability of ptDNA, MOC1 depletion in P. patens induced SDR-mediated ectopic recombination in mtDNA and disorganized cristae in mitochondria. Similar phenotypes in plastids and mitochondria were previously observed in mutants of plastid-targeted (RECA2) and mitochondrion-targeted (RECA1) recombinases, respectively. These results suggest that MOC1 functions in the double-strand break repair in which a recombinase generates HJs and MOC1 resolves HJs in mitochondria of some lineages of algae and plants in addition to plastids in algae and plants.
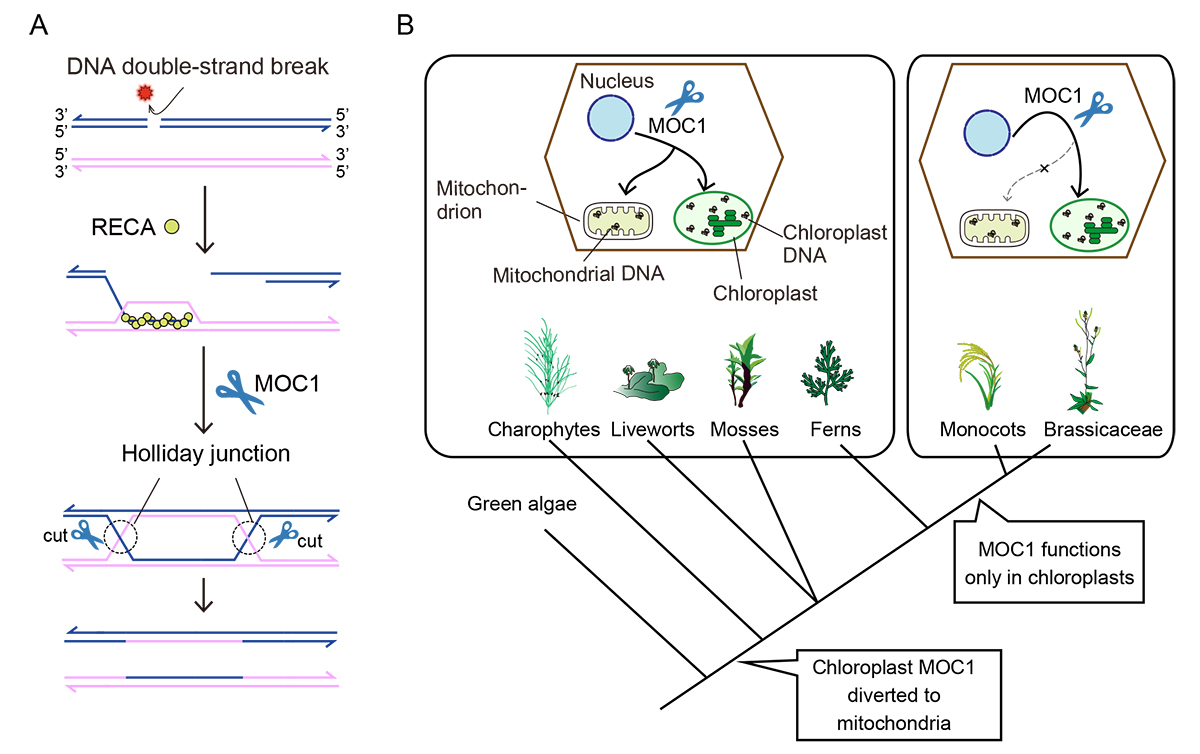
Figure: (A) Schematic diagram of double strand break repair that is mediated by a recombinase RECA and a Holliday junction resolvase MOC1.
(B) Evolutionary transition of MOC1-mediated organelle DNA maintenance in plants.
Neuronal Circuits That Control Rhythmic Pectoral Fin Movements in Zebrafish
Neuronal Circuits That Control Rhythmic Pectoral Fin Movements in Zebrafish
Yuto Uemura, Kagayaki Kato,Koichi Kawakami, Yukiko Kimura, Yoichi Oda, and Shin-ichi Higashijima
The Journal of Neuroscience 40, 6678-6690 (2020). DOI:10.1523/JNEUROSCI.1484-20.2020
Limbed vertebrates exhibit the basic form of locomotion, consisting of alternating activities of the flexor and extensor muscles within each limb coupled with left/right limb alternation. Zebrafish pectoral fin movements show the fundamental aspects of this basic movement: abductor/adductor alternation (corresponding to flexor/extensor alternation) and left/right fin alternation. Thus, zebrafish can serve as a good model to identify the neuronal networks of the central pattern generator (CPG) that controls rhythmic appendage movements. Here, we investigate neuronal circuits underlying rhythmic pectoral fin movements in larval zebrafish, using transgenic fish that specifically express GFP in abductor or adductor motor neurons (MNs) and candidate CPG neurons. First, we showed that spiking activities of abductor and adductor MNs are essentially alternating. Second, both abductor and adductor MNs receive rhythmic excitatory and inhibitory synaptic inputs in their active and inactive phases, respectively, indicating that the MN activities are controlled in a push-pull manner. Further, we obtained the evidence that dmrt3a-expressing commissural inhibitory neurons are involved in regulating the activities of abductor MNs: (1) strong inhibitory synaptic connections were found from dmrt3a neurons to abductor MNs; and (2) ablation of dmrt3a neurons shifted the spike timing of abductor MNs. Thus, in this simple system of abductor/adductor alternation, the last-order inhibitory inputs originating from the contralaterally located neurons play an important role in controlling the firing timings of MNs.
This study was conducted as collaboration with Professor Shin-ichi Higashijima at National Institute for Basic Biology. This study was partially supported by NBRP and NBRP/Fundamental Technologies Upgrading Program from the Japan Agency for Medical Research and Development.
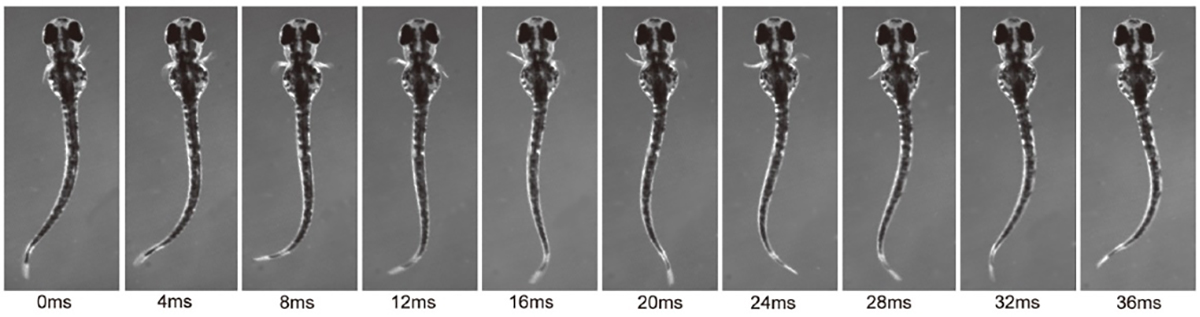
Figure: Rhythmic pectoral fin movements in 3 dpf larvae.
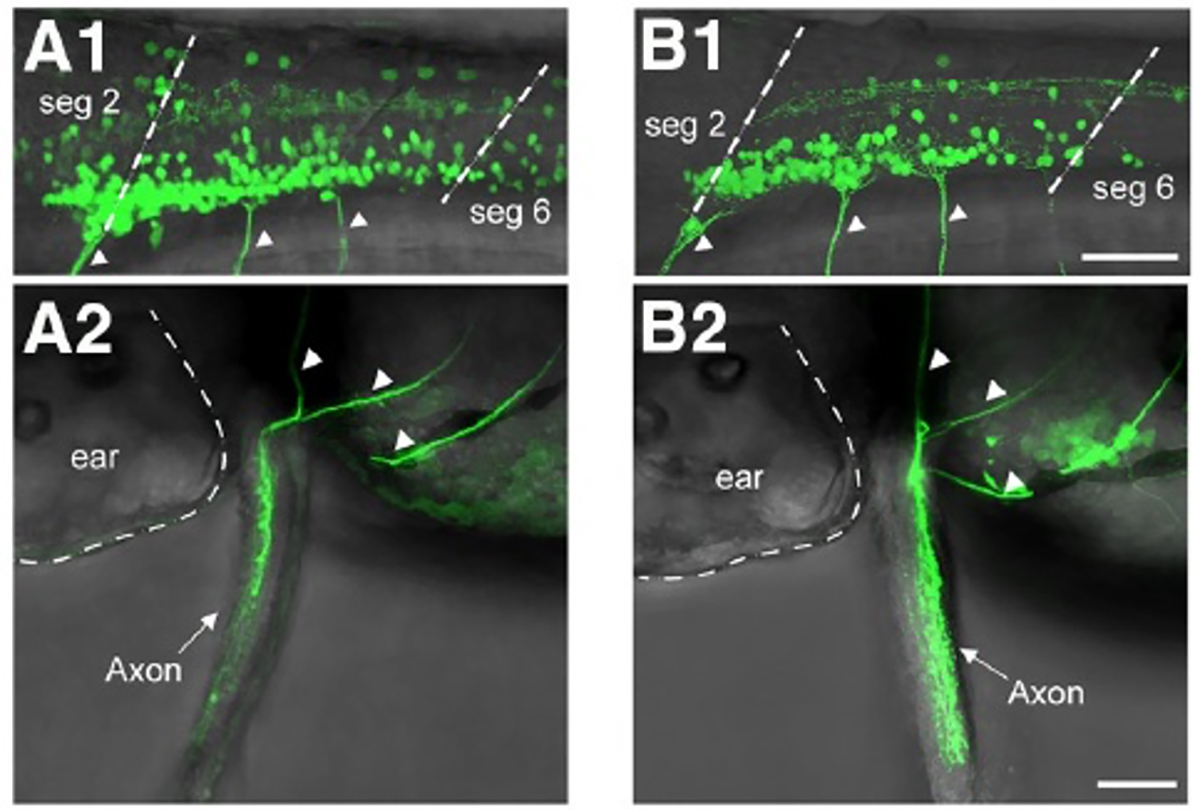
Figure: A1, A2 Transgenic fish that labeled abductor motor neurons.
B1, B2 Transgenic fish that labeled abductor motor neurons.
Mechanisms to silence mobile DNA within the genome
RNA interference-independent reprogramming of DNA methylation in Arabidopsis
Taiko Kim To, Yuichiro Nishizawa, Soichi Inagaki, Yoshiaki Tarutani, Sayaka Tominaga, Atsushi Toyoda, Asao Fujiyama, Frédéric Berger, Tetsuji Kakutani
Nature Plants 2020 November 30 DOI:10.1038/s41477-020-00810-z
DNA methylation is important for silencing transposable elements (TEs) in diverse eukaryotes including plants. In plant genomes, TEs are silenced by methylation of histone H3 lysine 9 (H3K9) and cytosines in both CG and non-CG contexts. The role of RNA interference (RNAi) in establishing TE-specific silent marks has been extensively studied, but the significance of RNAi-independent pathways remains largely unexplored. Here, we directly investigated transgenerational de novo DNA methylation of TEs after the loss of silent marks. Our analyses uncovered potent and precise RNAi-independent pathways for recovering non-CG methylation and H3K9 methylation in most TE genes (i.e. coding regions within TEs). Characterization of a subset of TE genes without the recovery revealed the impacts of H3K9 demethylation, replacement of histone H2A variants, and their interaction with CG methylation, together with feedback from transcription. These chromatin components are conserved among eukaryotes and may contribute to chromatin reprograming in a conserved manner.
Supported by Systems Functional Genetics Project of the Transdisciplinary Research Integration Center, ROIS.
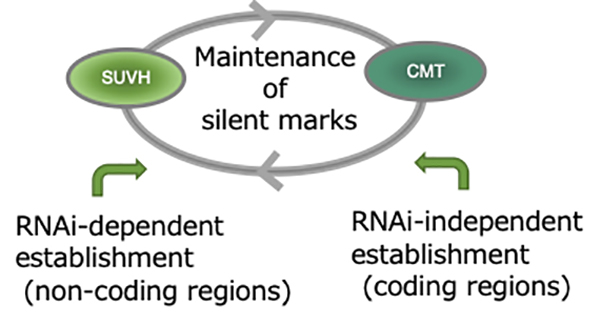
Figure: H3K9 and non-CG methylation can be maintained and inherited by mutual reinforcement. These marks are established by RNAi-dependent and -independent pathways. The RNAi-independent pathway mainly works in coding regions of TEs.















