Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
The Genetic Basis of Morphological Diversity in Domesticated Goldfish
Kawakami Group / Laboratory of Molecular and Developmental Biology
Comparative Genomics Laboratory
NOGUCHI, Hideki / Center for Genome Informatics(CGI) / Advanced Genomics Center
The Genetic Basis of Morphological Diversity in Domesticated Goldfish
Tetsuo Kon, Yoshihiro Omori, Kentaro Fukuta, Hironori Wada, Masakatsu Watanabe, Zelin Chen, Miki Iwasaki, Tappei Mishina, Shin-ichiro S. Matsuzaki, Daiki Yoshihara, Jumpei Arakawa, Koichi Kawakami, Atsushi Toyoda, Shawn M. Burgess, Hideki Noguchi, Takahisa Furukawa.
Current Biology 30, 1-15 (2020). DOI:10.1016/j.cub.2020.04.034
Although domesticated goldfish strains exhibit highly diversified phenotypes in morphology, the genetic basis underlying these phenotypes is poorly understood. Here, based on analysis of transposable elements in the allotetraploid goldfish genome, we found that its two subgenomes have evolved asymmetrically since a whole-genome duplication event in the ancestor of goldfish and common carp. We conducted whole- genome sequencing of 27 domesticated goldfish strains and wild goldfish. We identified more than 60 million genetic variations and established a population genetic structure of major goldfish strains. Genome-wide association studies and analysis of strain-specific variants revealed genetic loci associated with several goldfish phenotypes, including dorsal fin loss, long-tail, telescope-eye, albinism, and heart-shaped tail. Our results suggest that accumulated mutations in the asymmetrically evolved subgenomes led to generation of diverse phenotypes in the goldfish domestication history. This study is a key resource for understanding the genetic basis of phenotypic diversity among goldfish strains.
This work was carried out as collaboration between Osaka Univ and NIG (Lab of Molecular and Developmental Biology, Comparative Genomics Lab and Advanced Genomics Center) and supported by the NIG-JOINT program
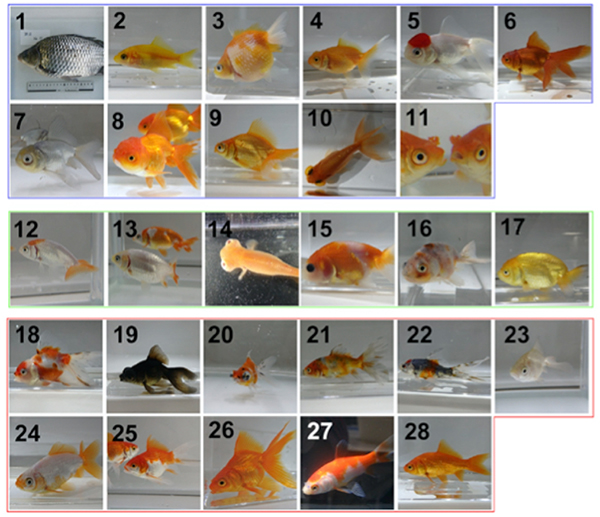
Figure: Wild type (1) and mutant (2-28) goldfish. Genome analysis revealed China (2-11), Ranchu (12-17) and Edo (18-28) groups.
Identification of a mechanism for the repair pathway choice after DNA double-strand break formation.
RIF1 Controls Replication Initiation and Homologous Recombination Repair in a Radiation Dose-Dependent Manner
Yuichiro Saito, Junya Kobayashi, Masato T. Kanemaki, Kenshi Komatsu
Journal of Cell Science 2020 May 20. DOI:10.1242/jcs.240036
Genomic DNA is challenged by various types of stresses. Ionizing radiation (IR) cut DNA, generating a DNA damage called a DNA double-strand break (DSB). DSB is repaired via two pathways; an error-prone, ‘non-homologous end joining (NHEJ) pathway’ or a precise, ‘homologous recombination repair (HRR) pathway’. To maintain genome integrity, it is important for cells to choose an appropriate pathway for repairing DSBs.
It is believed that human cells dominantly use error-prone NHEJ to repair DSB. This idea came from the observations using high doses of IR. However, it has not been known whether the predominant use of NHEJ is applicable to DSBs induced by a low dose of IR. To answer this question, we used a low or high dose of IR and monitored the repair pathway. We found that HRR played a prominent role to repair DSBs after a low dose of IR, and it was suppressed after a high dose of IR. These results show that human cells sense radiation dose and choose an appropriate repair pathway. Furthermore, we identified RIF1 as a critical factor that controls HRR activity in an IR-dose dependent manner. We expect that these results would provide basic understandings of how radiation kills cancer cells in radiation therapy.
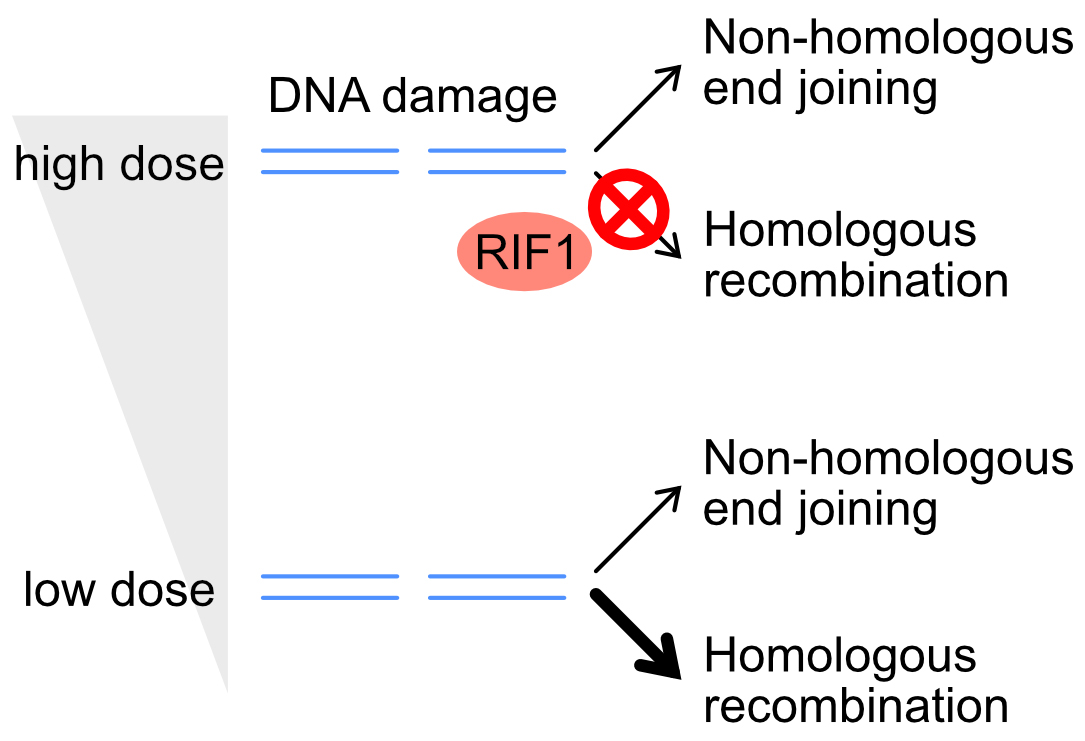
Figure: DNA double-strand break is repaired via the non-homologous end joining (NHEJ) or homologous recombination repair (HRR) pathway. RIF1 suppresses HRR only after a high dose of IR, directing cells towards using the NHEJ pathway.
- 1st author, Dr. Yuichiro Saito was introduced by the article “First person” in the journal.
Thievery leads to endosymbioses?
–a transient endosymbiosis in a kleptoplastic dinoflagellate
Changes in the transcriptome, ploidy, and optimal light intensity of a cryptomonad upon integration into a kleptoplastic dinoflagellate
Ryo Onuma, Shunsuke Hirooka, Yu Kanesaki, Takayuki Fujiwara, Hirofumi Yoshikawa, Shin-ya Miyagishima
The ISME Journal (2020) DOI:10.1038/s41396-020-0693-4
Link to “Behind the Paper” the Nature Research Microbiology Community
Endosymbiosis of unicellular eukaryotic algae into previously nonphotosynthetic eukaryotes has established chloroplasts in several eukaryotic lineages. Additionally, certain unicellular organisms in several different lineages ingest algae and utilize them as temporal chloroplasts (kleptoplasts) for weeks to months before digesting them. Among these organisms, the dinoflagellate Nusuttodinium aeruginosum ingests the cryptomonad Chroomonas sp. and enlarges the kleptoplast with the aid of the cryptomonad nucleus. To understand how the cryptomonad nucleus is remodeled in the dinoflagellate, here we examined changes in the transcriptome and ploidy of the ingested nucleus. We show that, after ingestion, genes involved in metabolism, translation, and DNA replication are upregulated while those involved in sensory systems and cell motility are downregulated. In the dinoflagellate cell, the cryptomonad nucleus undergoes polyploidization that correlates with an increase in the mRNA levels of upregulated genes. In addition, the ingested nucleus almost loses transcriptional responses to light. Because polyploidization and loss of transcriptional regulation are also known to have occurred during the establishment of endosymbiotic organelles, these changes are probably a common trend in endosymbiotic evolution. Furthermore, we show that the kleptoplast and dinoflagellate are more susceptible to high light than the free-living cryptomonad but that the ingested nucleus reduces this damage.
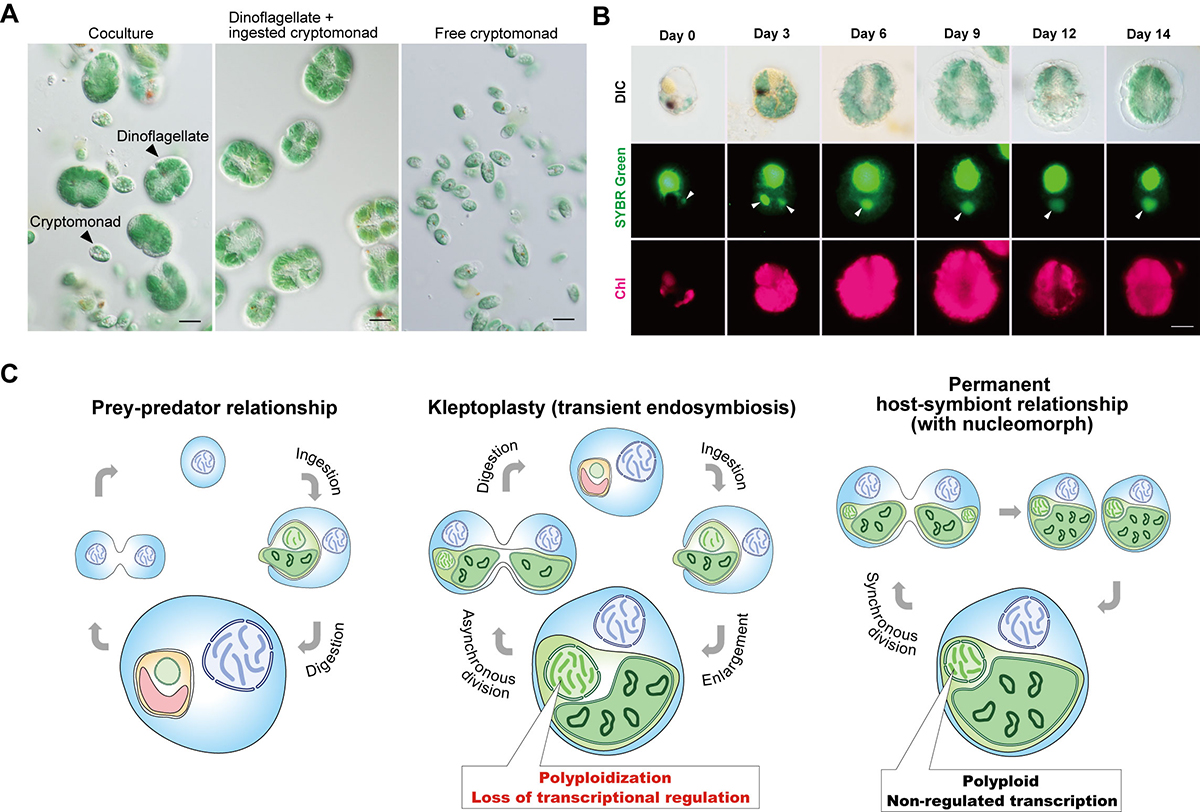
Figure: Kleptoplasty in Nusuttodinium aeruginosum.
(A) Micrographs showing the coculture of N. aeruginosum cells with Chroomonas sp.-derived kleptoplasts and free Chroomonas sp. (left). From the coculture, N. aeruginosum cells with the kleptoplasts (middle) were separated from free Chroomonas sp. (right) by filtration. Scale bar = 10 μm.
(B) Micrographs showing the change in the nucleus and kleptoplast derived from Chroomonas sp. in N. aeruginosum cells. Cells were stained with SYBR Green. Images of differential interference contrast (DIC), SYBR Green staining, and kleptoplast red fluorescence (Chl) are shown. The arrowhead indicates a nucleus derived from Chroomonas sp. The cell in the image from day 3 ingested two nuclei. Scale bar = 10 μm.
(C) Schematic comparison of prey–predator, transient endosymbiotic (kleptoplasty), and permanent endosymbiotic relationships. Predators start digesting algae immediately after phagocytic ingestion of algae (left). Kleptoplastic species retain the ingested algae for days to weeks in the cell before digesting them in some cases, including that of N. aeruginosum, with the nucleus of the algae (middle). The present study shows that the ingested nucleus is polyploidized in the host cell, which increases mRNA levels. The ingested nucleus almost loses transcriptional regulation. These changes were also common to the course of establishment of nucleomorph and chloroplast (also other types of plastids) as obligate endosymbionts in eukaryotes (right).















