Transcription factor triggering small RNA biogenesis in rice anthers
Experimental Farm / Nonomura Group
EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum.
Seijiro Ono, Hua Liu, Katsutoshi Tsuda, Eigo Fukai, Keisuke Tanaka, Takuji Sasaki, Ken-Ichi Nonomura.
PLOS Genetics, 14 (2), e1007238, (2018) DOI:10.1371/journal.pgen.1007238
Small RNA plays important roles in development, anti-viral defense and etc., mediated by transcriptional and post-transcriptional of repression of gene expressions. Here, we revealed that the EAT1 transcription factor triggers the biosynthesis of 24-nt small RNAs during meiosis in rice anther tapetum. EAT1 promoted the transcription of long precursor RNAs, as small RNA precursors, and DCL5 gene, required for small RNA processing (Fig. 1).
This study demonstrated a possibility that a subset of EAT1-dependent 24-nt phasiRNAs are transferred from tapetal cells to adjoining meiocytes, from the analysis of MEL1, the Argonaute protein specifically expressed in meiocytes (Fig. 1C). This finding strongly suggests that the reproductive phasiRNAs may act as a mediator of orchestrated development of germ cells and surrounding somatic cells.
This work is a collaboration with Tokyo University of Agriculture and Niigata University.
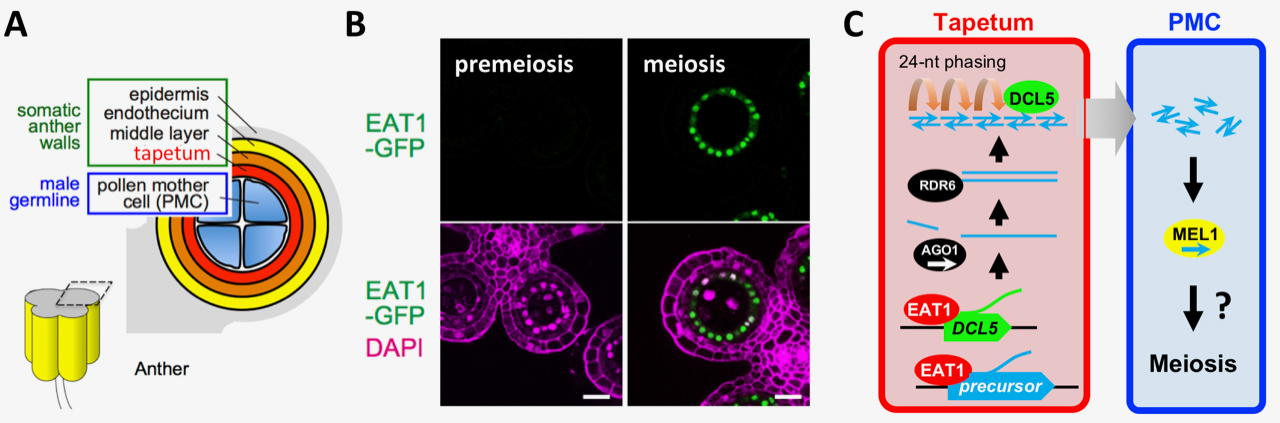
Fig. 1. EAT1 transcription factor promotes the biosynthesis of 24-nt meiotic small RNA (sRNA) in rice anther tapetum
(A) A cross section of an anther lobe. Tapetum is adjoining to male meiocytes (PMCs). (B) EAT1 (green) accumulates in tapetal cell nuclei at meiosis (right), but not at premeiosis (left). Bar=20µm. (C) EAT1 activates transcription of sRNA precursors and DCL5 gene in tapetum. After primary processing and double-strandization, precursor RNAs are sliced into 24 nt by DCL5. MEL1 binding to a subset of EAT1-dependent sRNA suggests intercellular mobilization of meiotic sRNA.















