Release of an upgraded NIG-Mouse Genome Database, “NIG_MoG2”, to the public
Mammalian Genetics Laboratory / Shiroishi Group
Genetic Informatics Laboratory / Kawamoto Group
Commonly used laboratory mouse strains have mosaic genomes, which are derived predominantly from the west European subspecies with the remaining sequences derived from the Japanese subspecies. National Institute of Genetics (NIG) has established a series of experimental mouse strains named “Mishima Battery”. It consists of ten inbred strains originated from four mouse subspecies including the Japanese subspecies, which are widely distributed in the world (Fig. 1). Most of these strains have very remote genetic status from the commonly used laboratory mouse strains such as C57BL/6J (B6), and show very unique phenotypes. Genomic polymorphisms between the laboratory strains and the Japanese subspecies-derived strains are widely used in studies of epigenetics as allele-specific markers. The mouse strains in the “Mishima Battery” are now distributed to the research community via RIKEN BioResource center and NIG, and contribute to broad range of life science. NIG has also established B6-ChrNMSM consomic strains, in which every chromosome of B6 is substituted by corresponding chromosome of Japanese wild mouse-derived MSM/Ms strain, a member of “Mishima Battery”. Using a full panel of the consomic strains, researchers are able to identify chromosomes that control complex traits and diseases. NIG provides these consomic strains to the research community.
Our recent genome-resequencing project of “Mishima Battery” revealed over forty millions of SNPs due to large inter-subspecific genetic distance. Here, we release an upgraded version of the NIG Mouse Genome database named “NIG_MoG2” (http://molossinus.lab.nig.ac.jp/mog2/). NIG_MoG2 primarily comprises the whole genome sequence data of the “Mishima Battery” (Fig. 2), providing visualized genome polymorphism information with single-nucleotide polymorphisms and short insertions/deletions in the genomes of the “Mishima Battery”. It allows users, especially for wet-lab biologists, to intuitively recognize inter-subspecific genome divergence on the browser. NIG_MoG2 is thus a valuable navigator for exploring mouse genome polymorphisms.
This project was supported by NIG Advanced Genomics Project (Next Generation Genome-Research-Hub Project), “Genome Information Upgrading Program” of National BioResource Project (NBRP) from Japan Agency for Medical Research and Development (AMED). The project was performed as a mission of the Genetic Resource Center of NIG.
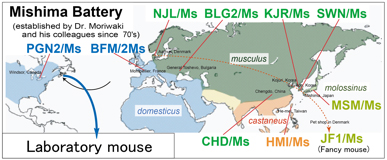
Fig. 1. Geographic distribution of mouse subspecificies, and origins of the “Mishima Battery”. Names of the strains are indicated in colored and bold prints.
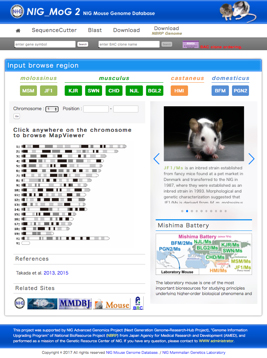
Fig. 2. Top page of the NIG_MoG2 (http://molossinus.lab.nig.ac.jp/mog2).















