Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis.
Division of Agricultural Genetics • Kakutani Group
Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis.
Tasuku Ito, Yoshiaki Tarutani, Taiko Kim To, Mohamed Kassam, Evelyne Duvernois-Berthet, Sandra Cortijo, Kazuya Takashima, Hidetoshi Saze, Atsushi Toyoda, Asao Fujiyama, Vincent Colot, Tetsuji Kakutani PLoS Genetics Published: April 22, 2015 DOI:10.1371/journal.pgen.1005154DNA methylation is important for controlling activity of transposable elements and genes. An intriguing feature of DNA methylation in plants is that its pattern can be inherited over multiple generations at high fidelity in a Mendelian manner. However, mechanisms controlling the trans-generational DNA methylation dynamics are largely unknown. Arabidopsis mutants of a chromatin remodeler gene DDM1 (Decrease in DNA Methylation 1) show drastic reduction of DNA methylation in transposons and repeats, and also show progressive changes in developmental phenotypes during propagation through self-pollination. We now show using whole genome DNA methylation sequencing that upon repeated selfing, the ddm1 mutation induces an ectopic accumulation of DNA methylation at hundreds of loci. Remarkably, even in the wild type background, the analogous de novo increase of DNA methylation can be induced in trans by chromosomes with reduced DNA methylation. Collectively, our findings support a model to explain the transgenerational DNA methylation redistribution by genome-wide negative feedback, which should be important for balanced differentiation of DNA methylation states within the genome. This work is a collaboration with Fujiyama lab supported by Systems Functional Genetics Project of the Transdisciplinary Research Integration Center, ROIS.
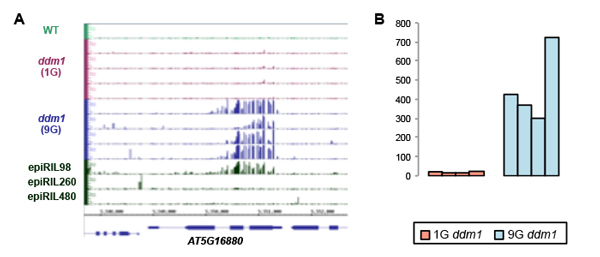
(A) Ectopic increase of cytosine methylation in the ddm1 mutant plants. The methylation accumulated in the 9th generation of ddm1 mutant (9G), but not in the 1st generation (1G). A line with large amount of chromosomes from ddm1 (epiRIL98) introduced into DDM1 wild type background also showed the ectopic DNA methylation. (B) Similar ectopic DNA methylation can be seen at hundreds of loci specifically in the 9G ddm1 mutant plants. The value shows number of genes with ectopic methylation in each of four 9G and four 1G ddm1 lines.















