Archive
- Home
- February 2026
- January 2026
- December 2025
- November 2025
- October 2025
- September 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- March 2025
- February 2025
- January 2025
- December 2024
- November 2024
- October 2024
- September 2024
- August 2024
- July 2024
- June 2024
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014
- January 2014
- December 2013
- November 2013
- October 2013
- September 2013
- August 2013
- July 2013
- June 2013
- May 2013
- April 2013
- March 2013
- February 2013
- January 2013
- December 2012
- November 2012
- October 2012
- September 2012
- August 2012
- July 2012
- June 2012
- May 2012
- April 2012
- March 2012
- February 2012
- January 2012
- December 2011
- November 2011
- October 2011
- September 2011
- August 2011
- July 2011
- June 2011
- May 2011
- April 2011
- March 2011
- February 2011
- January 2011
- December 2010
- November 2010
- October 2010
- September 2010
- August 2010
- July 2010
- June 2010
- May 2010
- April 2010
- March 2010
- February 2010
- December 2009
- November 2009
- October 2009
- September 2009
- August 2009
- July 2009
- June 2009
- May 2009
- April 2009
- March 2009
- February 2009
- January 2009
- December 2008
- November 2008
- October 2008
- September 2008
- July 2008
- June 2008
- May 2008
- April 2008
- March 2008
- February 2008
- January 2008
- December 2007
- November 2007
- October 2007
- September 2007
- August 2007
- July 2007
- June 2007
- March 2007
- February 2007
- January 2007
- December 2006
- November 2006
- October 2006
- September 2006
- July 2006
- June 2006
- May 2006
- April 2006
- January 2006
- June 2005
- May 2005
- April 2005
- February 2005
- December 2004
- November 2004
- October 2004
- August 2004
- July 2004
- June 2004
- May 2004
- February 2004
- January 2004
- December 2003
- August 2003
- July 2003
- June 2003
- May 2003
- March 2003
- January 2003
- December 2002
- October 2002
- May 2002
- April 2002
- February 2002
- January 2002
- August 2001
- May 2001
- April 2001
- February 2001
- August 2000
- July 2000
Improvement of Pyrrole−Imidazole Polyamide Probes Targeting Human Telomeres
Biological Macromolecules Laboratory • Maeshima Group
(i) Structural Evaluation of Tandem Hairpin Pyrrole–Imidazole Polyam-ides Recognizing Human Telomeres
Hirata, A., Nokihara, K., Kawamoto, Y., Bando, T., Sasaki, A., Ide, S., Maeshima, K., Kasama, T., and Sugiyama, H. Journal of the American Chemical Society (JACS), July 18, 2014. DOI: 10.1021/ja506058e
(ii) Tandem Trimer Pyrrole-Imidazole Polyamide Probes Targeting 18 Base Pairs in Human Telomere Sequences
Kawamoto, Y., Sasaki, A., Hashiya, K., Ide, S., Bando, T. *, Maeshima, K. *, and Sugiyama, H.**co-corresponding authors
Chemical Science, January 20, 2015. DOI: 10.1039/C4SC03755C
Pyrrole-Imidazole (PI) polyamides bind to the minor groove of DNA in a sequence-specific manner. Our previous studies have demonstrated that a synthesized tandem hairpin PI polyamide (TH59) can target human telomere sequences (TTAGGG)n under mild conditions and can serve as a new probe for studying human telomeres. In the recently published two papers, we have improved specificity of the tandem hairpin PI polyamide by optimization of the connecting region (hinge region) of the tandem hairpin parts [HPTH59-b in Fig. A and paper (i)] and by introducing additional hairpin part [TT59 in Fig. B and paper (ii)]. The HPTH59-b and TT59 have higher specificity to recognize human telomeric repeats than the previous ones and stain telomeres in chemically fixed human cells with lower background signal.
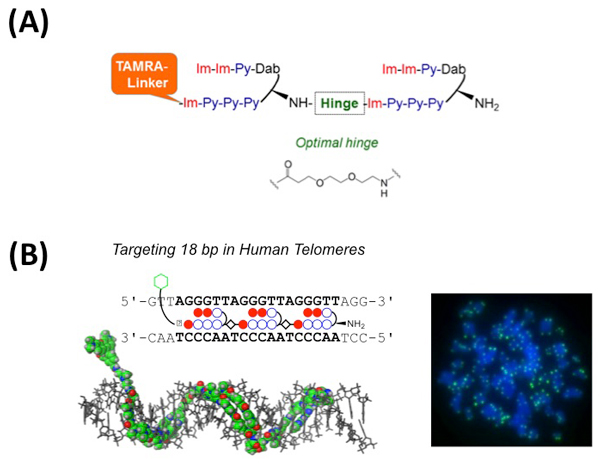
(A) Schematic structure of HPTH59-b. The optimized chemical structure of hinge segment is shown.
(B) Left, schematic representation of the tandem trimer PI polyamide TT59, which binds to 18bp of the human telomere sequence. Right, the telomere regions of chromosome ends are labeled with TT59 (green). DNA stain (blue).
Professor emeritus Tomoko Ohta awarded the Crafoord Prize in Biosciences
The Royal Swedish Academy of Sciences has awarded the 2015 Crafoord Prize in Biosciences to Professor emeritus Tomoko Ohta and Professor emeritus Richard Lewontin for their pioneering work in understanding genetic variation and evolution. The award ceremony will take place on May 5-7 in Stockholm, Sweden. The Crafoord Prize in astronomy and mathematics, biosciences, geosciences or polyarthritis research is awarded annually according to a rotating scheme. These disciplines are chosen to complement those for which the Nobel Prizes are awarded.
This year’s prize recognizes Professor Ohta’s fundamental contributions to the understanding of genetic polymorphism conducted at NIG. Her work, including “Slightly Deleterious Mutant Substitutions in Evolution” published by Nature in 1973 proposed “near neutrality” as a important factor for understanding both variation within a population and differences among species. Weak selection is now considered central to understanding genome evolution and her ideas have permeated thinking in biomedical research and systems biology as well as in comparative genomics.
The physical size of transcription factors is key to transcriptional regulation in chromatin domains
Biological Macromolecules Laboratory • Maeshima Group
The physical size of transcription factors is key to transcriptional regulation in chromatin domains
Kazuhiro Maeshima, Kazunari Kaizu, Sachiko Tamura, Tadasu Nozaki, Tetsuro Kokubo, and Koichi TakahashiJournal of Physics: Condensed Matter, 27, 064116 (10 pp), 2015. DOI: 10.1088/0953-8984/27/6/064116
Genetic information, which is stored in the long strand of genomic DNA as chromatin, must be scanned and read out by various transcription factors. First, gene-specific transcription factors, which are relatively small (~50 kDa), scan the genome and bind regulatory elements. Such factors then recruit general transcription factors, Mediators, RNA polymerases, nucleosome remodellers, and histone modifiers, most of which are large protein complexes of 1–3 MDa in size. Here, we propose a new model for the functional significance of the size of transcription factors (or complexes) for gene regulation of chromatin domains. Recent findings suggest that chromatin consists of irregularly folded nucleosome fibres (10 nm fibres) and forms numerous condensed domains (e.g., topologically associating domains)(blue balls in Fig). Although the flexibility and dynamics of chromatin allow repositioning of genes within the condensed domains, the size exclusion effect of the domain may limit accessibility of DNA sequences by transcription factors. We used Monte Carlo computer simulation to determine the physical size limit of transcription factors that can enter condensed chromatin domains. Small gene-specific transcription factors can penetrate into the chromatin domains and search their target sequences, whereas large transcription complexes cannot enter the domain (Fig a). Due to this property, once a large complex binds its target site via gene-specific factors it can act as a “buoy” to keep the target region on the surface of the condensed domain (Fig b) and maintain transcriptional competency (Fig c). This size-dependent specialisation of target-scanning and surface-tethering functions could provide novel insight into the mechanisms of various DNA transactions, such as DNA replication and repair/recombination.
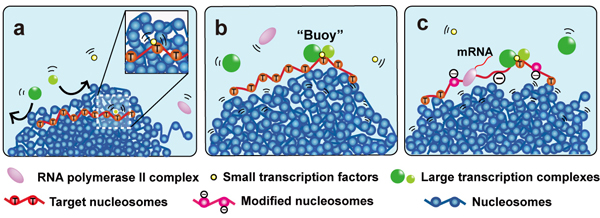
“Buoy” Model for Transcriptional Regulation
Condensed chromatin domains are shown with blue nucleosomes.
(a) The small transcription factors in yellow can move in the condensed chromatin domain but not large transcription complexes (in green). (b) The small transcription factor bound to the target region (red nucleosomes) and can recruit large transcription complexes when it is relocated on the domain surface. Binding of large transcription complexes (green) keeps the transcriptional region (red nucleosomes) on the domain surface like a ‘buoy’. (c) With other large complexes such as nucleosome remodeler or histone modifier, stable transcription is maintained.
Novel mechanism of DNA duplex unwinding by a single protein
超分子構造研究室・白木原研究室
Structural basis for replication origin unwinding by an initiator-primase of plasmid ColE2-P9: Duplex DNA unwinding by a single protein
Hiroshi Itou, Masaru Yagura, Yasuo Shirakihara, and Tateo ItohJournal of Biological Chemistry, 2015 Feb 6;290(6):3601-3611. DOI: 10.1074/jbc.M114.595645
Duplex DNA is generally unwound by protein oligomers prior to replication. The Rep protein of plasmid ColE2-P9 is an essential initiator for plasmid DNA replication. This protein binds the replication origin (Ori) in a sequence specific manner as a monomer and unwinds DNA. We present the crystal structure of the DNA-binding domain of Rep (E2Rep-DBD) in complex with Ori DNA. The structure unveils the basis for Ori specific recognition by the E2Rep-DBD and also reveals that it unwinds DNA by the concerted actions of its three contiguous structural modules. The structure also shows that the functionally unknown PriCT domain, which forms a compact module, plays a central role in DNA unwinding. The conservation of the PriCT domain in the C-termini of some archaeo-eukaryotic primases, indicates that it likely plays a similar role in these proteins. Thus, this is the first report providing the structural basis for the functional importance of the conserved PriCT domain and also reveals a novel mechanism for DNA unwinding by a single protein.
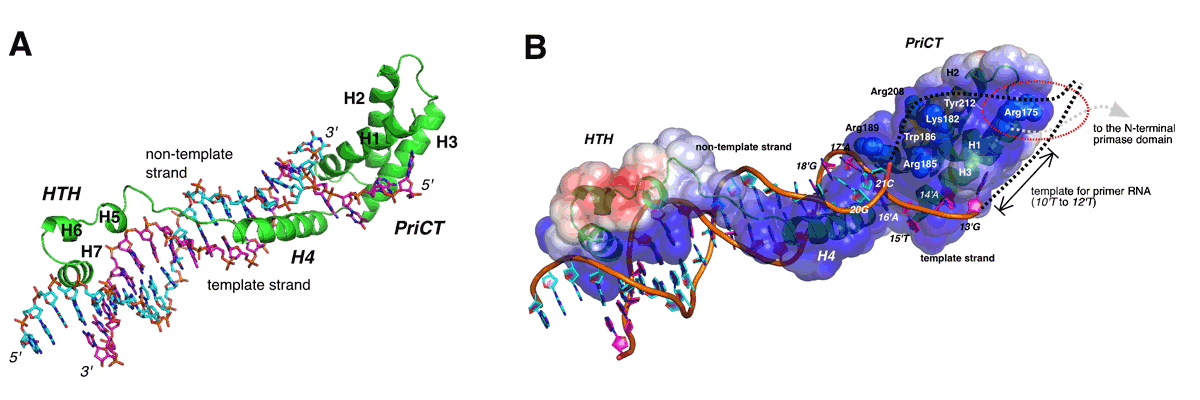
(A) The ribbon model of the complex consists of E2Rep-DBD and Ori DNA. Elongated fold of E2Rep-DBD enables binding specificity and affinity enough to unwind duplex DNA.
(B) Model for unwinding of the complete Ori by Rep. The PriCT-module plays a central role in unwinding of duplex DNA and stabilizing the single-stranded DNA.















