Chromatin foundation for the kinetochore machinery
Division of Molecular Genetics • Fukagawa Group
The centromere: chromatin foundation for the kinetochore machinery
Tatsuo Fukagawa, and William C. Earnshaw Dev. Cell 30,496-508,2014; DOI:10.1016/j.devcel.2014.08.016Since discovery of the centromere-specific histone H3 variant CENP-A, centromeres have come to be defined as chromatin structures that establish the assembly site for the complex kinetochore machinery. In most organisms, centromere activity is defined epigenetically, rather than by specific DNA sequences. In this review, Fukagawa (Dep. Mol. Genet.) and Earnshaw (ex visiting professor of NIG, Edinburgh Univ) describe selected classic work and recent progress in studies of centromeric chromatin with a focus on vertebrates. We consider possible roles for repetitive DNA sequences found at most centromeres, chromatin factors and modifications that assemble and activate CENP-A chromatin for kinetochore assembly, and use of artificial chromosomes and kinetochores to study centromere function.
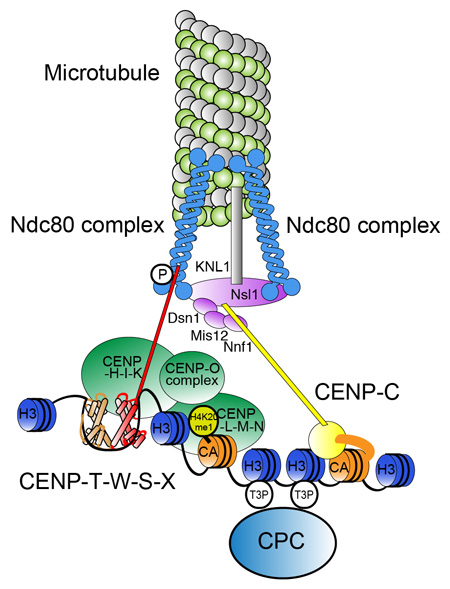
Molecular architecture of natural kinetochores. Centromere chromatin including is crucial for centromere specification and kinetochore assembly at natural centromeres. At the base of the structure are CENP-A containing nucleosomes, centromere specific H3 nucleosomes, and a CENP-T-W-S-X nucleosome-like structure in centromere chromatin. Centromere-specific chromatin structure is established by coordination of these components. CCAN proteins assemble on the centromeric chromatin and the microtubule-binding complex is subsequently recruited to assemble the functional kinetochore.















