Mechanism for transcribing Arabidopsis genes containing intragenic heterochromatin
Division of Agricultural Genetics • Kakutani Group
Hidetoshi Saze, Junko Kitayama, Kazuya Takashima, Saori Miura, Yoshiko Harukawa, Tasuku Ito and Tetsuji Kakutani
Nature Communications, 4, Article number:2301 doi:10.1038/ncomms3301
Genomes of vertebrates and plants contain a substantial number of transposable elements (TEs), which are silenced by repressive epigenetic modifications, such as cytosine methylation and methylation of lysine 9 of histone H3. These modifications are essential for formation of inactive chromatin structures called heterochromatin. A potential complication is that active cellular genes sometimes contain TEs within their transcribed regions.
In this study, we show that heterochromatic epigenetic modifications are commonly found within actively transcribed gene units in both the Arabidopsis and rice genomes. We further show that in Arabidopsis, full-length transcription of genes with intragenic heterochromatin, most of which is formed by TE insertions, requires IBM2 (Increase in Bonsai Methylation 2), a protein with a Bromo-Adjacent Homology (BAH) domain and an RNA recognition motif (RRM). Our results reveal a novel epigenetic mechanism that masks effects of genetic variations created by TE insertions, allowing evolution of complex genomes with heterochromatic domains having diverse functions. (This work is a collaboration with Saze lab in OIST.)
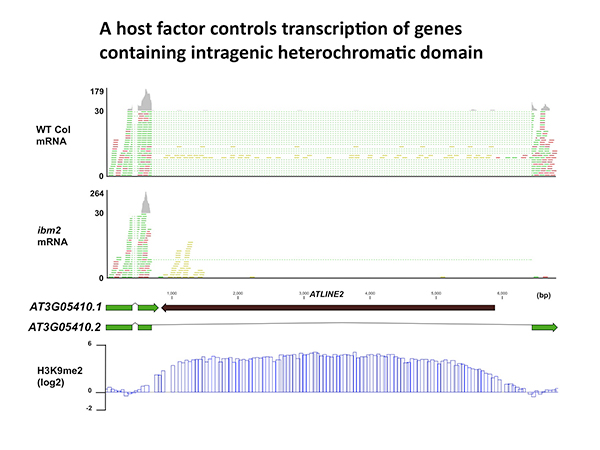
Full-length transcription of the gene containing heterochromatin is impaired in ibm2 mutant. mRNA reads are indicated with green and red. Reads with dotted lines are mapped across exon-exon boundaries. Note that the inserted TE (ATLINE2) is associated with heterochromatic H3K9 methylation (bottom).















