The physical size of transcription factors is key to transcriptional regulation in chromatin domains
Biological Macromolecules Laboratory • Maeshima Group
The physical size of transcription factors is key to transcriptional regulation in chromatin domains
Kazuhiro Maeshima, Kazunari Kaizu, Sachiko Tamura, Tadasu Nozaki, Tetsuro Kokubo, and Koichi TakahashiJournal of Physics: Condensed Matter, 27, 064116 (10 pp), 2015. DOI: 10.1088/0953-8984/27/6/064116
Genetic information, which is stored in the long strand of genomic DNA as chromatin, must be scanned and read out by various transcription factors. First, gene-specific transcription factors, which are relatively small (~50 kDa), scan the genome and bind regulatory elements. Such factors then recruit general transcription factors, Mediators, RNA polymerases, nucleosome remodellers, and histone modifiers, most of which are large protein complexes of 1–3 MDa in size. Here, we propose a new model for the functional significance of the size of transcription factors (or complexes) for gene regulation of chromatin domains. Recent findings suggest that chromatin consists of irregularly folded nucleosome fibres (10 nm fibres) and forms numerous condensed domains (e.g., topologically associating domains)(blue balls in Fig). Although the flexibility and dynamics of chromatin allow repositioning of genes within the condensed domains, the size exclusion effect of the domain may limit accessibility of DNA sequences by transcription factors. We used Monte Carlo computer simulation to determine the physical size limit of transcription factors that can enter condensed chromatin domains. Small gene-specific transcription factors can penetrate into the chromatin domains and search their target sequences, whereas large transcription complexes cannot enter the domain (Fig a). Due to this property, once a large complex binds its target site via gene-specific factors it can act as a “buoy” to keep the target region on the surface of the condensed domain (Fig b) and maintain transcriptional competency (Fig c). This size-dependent specialisation of target-scanning and surface-tethering functions could provide novel insight into the mechanisms of various DNA transactions, such as DNA replication and repair/recombination.
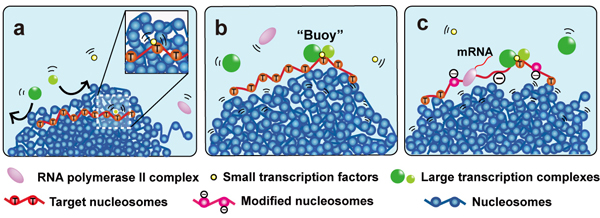
“Buoy” Model for Transcriptional Regulation
Condensed chromatin domains are shown with blue nucleosomes.
(a) The small transcription factors in yellow can move in the condensed chromatin domain but not large transcription complexes (in green). (b) The small transcription factor bound to the target region (red nucleosomes) and can recruit large transcription complexes when it is relocated on the domain surface. Binding of large transcription complexes (green) keeps the transcriptional region (red nucleosomes) on the domain surface like a ‘buoy’. (c) With other large complexes such as nucleosome remodeler or histone modifier, stable transcription is maintained.















