Akashi Group • Evolutionary Genetics Laboratory
Population genetics and genome evolution
Faculty
▶ Lab HP
Research Summary
Our group studies mechanisms of genome evolution with a focus on global adaptations and the dynamics of weak selection. We integrate functional genomics data and evolutionary models to motivate our analyses. Current topics include:
(1) phenotypic bases and population genetics of selection on synonymous codon usage
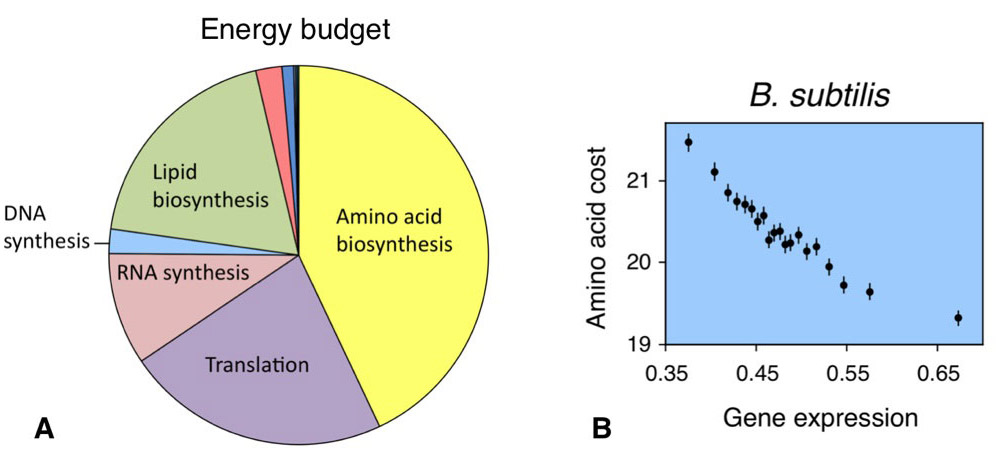
(2) amino acid composition and biosynthetic constraints in protein evolution
(3) protein structure/function and modes of slightly deleterious and adaptive evolution.
Selected Publications
Matsumoto T, John A, Baeza-Centurion P, Li B, Akashi H. Codon Usage Selection Can Bias Estimation of the Fraction of Adaptive Amino Acid Fixations. Mol Biol Evol. 2016 Jun;33(6):1580-9.
Matsumoto T, Akashi H, Yang Z. Evaluation of Ancestral Sequence Reconstruction Methods to Infer Nonstationary Patterns of Nucleotide Substitution. Genetics. 2015 Jul;200(3):873-90.
Akashi H, Osada N, Ohta T. Weak selection and protein evolution. Genetics. 2012 Sep;192(1):15-31.
Matsumoto T, Akashi H. Distinguishing Among Evolutionary Forces Acting on Genome-Wide Base Composition: Computer Simulation Analysis of Approximate Methods for Inferring Site Frequency Spectra of Derived Mutations. G3 (Bethesda). 2018 May 4;8(5):1755-1769.
















