Where are the replication initiation zones located in mammalian genome? (Review paper)
Kanemaki Group / Molecular Cell Engineering Laboratory
Replication initiation sites and zones in the mammalian genome: Where are they located and how are they defined?
Xiaoxuan Zhu, Masato T. Kanemaki*
*Corresponding author
DNA Repiar (2024) 141, 103713 DOI:10.1016/j.dnarep.2024.103713
The question of how genomic DNA is replicated is one of the key themes in molecular biology, as abnormalities in DNA replication are related to genome instability, genetic disorders, cell death, and cancer. Therefore, DNA replication has been extensively studied over the years using Escherichia coli and budding yeast as model organisms. In these unicellular models, DNA replication starts from genomic regions with consensus DNA sequences. On the other hand, in mammalian cells, including humans, consensus sequences appear to be absent, and it is still not well understood where DNA replication begins and how are they defined in the genome. Hence, this review paper compiles and compares the results of large-scale high-throughput sequencing-based explorations for mapping initiation sites/zones reported so far. Furthermore, it highlights the similarities and differences in the DNA replication mechanisms between budding yeast and mammalian cells, presents a hypothetical mechanism for how replication initiation zones are defined in mammalian cells, and discusses the future directions and challenges in DNA replication research.
The first author, Dr Xiaoxuan Zhu, has been supported as an NIG post-doctoral fellow.
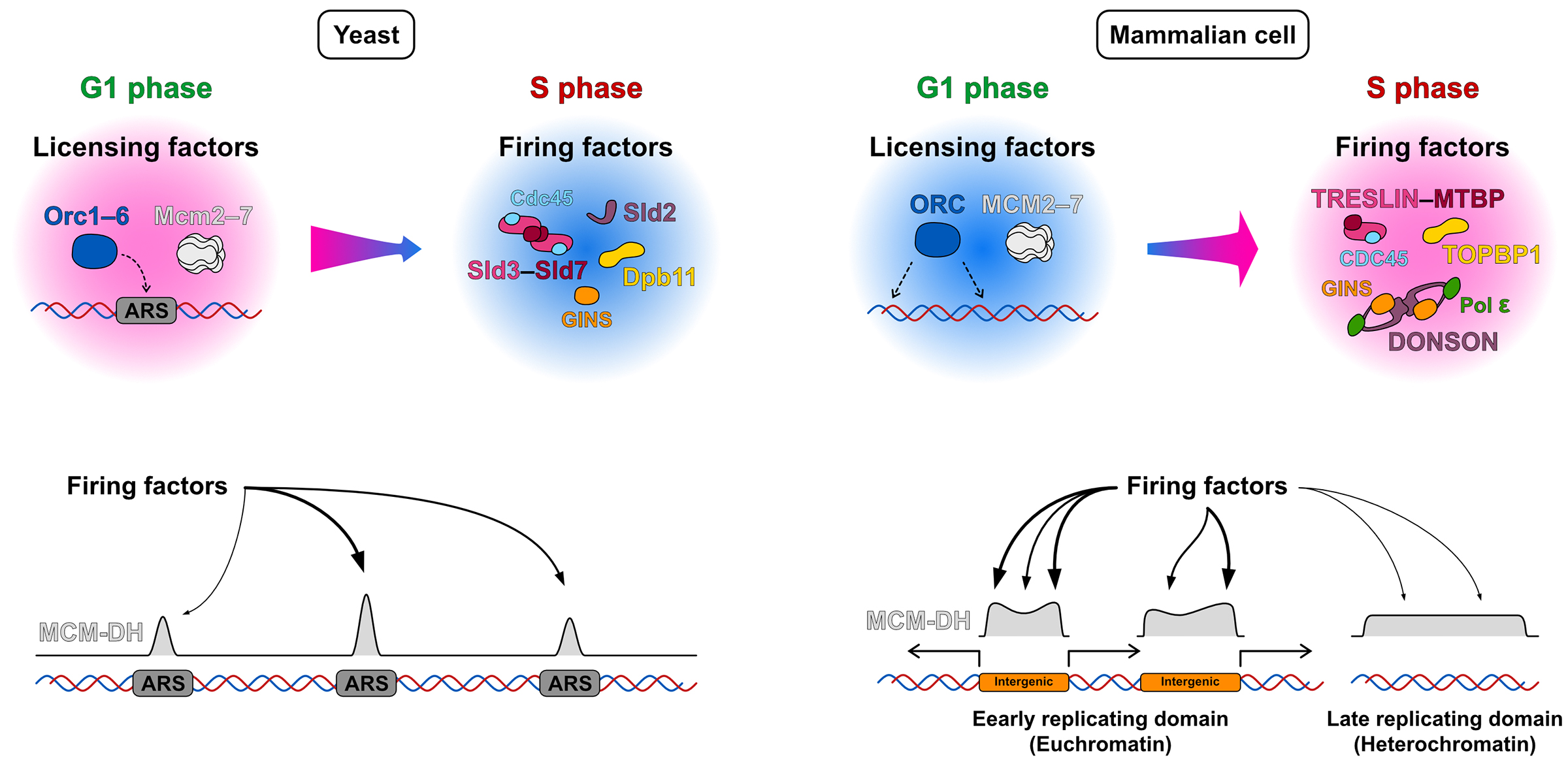
Figure: Differences in the mechanisms of replication initiation sites/zones selection between budding yeast and mammalian cells: In yeast, licensing factors accumulate at ARS (autonomously replicating sequence) sites containing a consensus sequence, leading to replication initiation at these ARS sites. In contrast, in mammalian cells, licensing factors are widely distributed across the genome. It is thought that the subsequent accumulation of firing factors at specific locations is crucial for forming initiation zones.















