Mechanism to coordinate overlapping transcription
Chromatin-based mechanisms to coordinate convergent overlapping transcription.
Soichi Inagaki, Mayumi Takahashi, Kazuya Takashima, Satoyo Oya, Tetsuji Kakutani.
Nature Plants 2021 March 1 DOI:10.1038/s41477-021-00868-3
In addition to coding genes, non-coding regions are often transcribed to make the transcription pattern overlapped. How organisms deal with overlapping transcription between neighboring genes remain unanswered. In this study we used a flowering plant Arabidopsis thaliana with a compact and gene-dense genome and found a mechanism to coordinate overlapping transcription in the opposite directions. We found that a histone demethylase protein FLD (flowering locus D) specifically controls genes with overlapping transcription by removing monomethylation of histone H3 lysine 4 (H3K4me1). We also showed that this regulation by FLD is linked to the mechanism to prevent the supercoiling or entanglement of DNA, which is crucial to maintain the genome. Interestingly, this global mechanism to coordinate overlapping transcription is utilized by plants to know the timing when they make flowers by adjusting the gene expression epigenetically in response to temperature. The findings of this study would lead to an understanding of the mechanism to cope with the crowded genome to properly regulate gene expression and respond to environment.
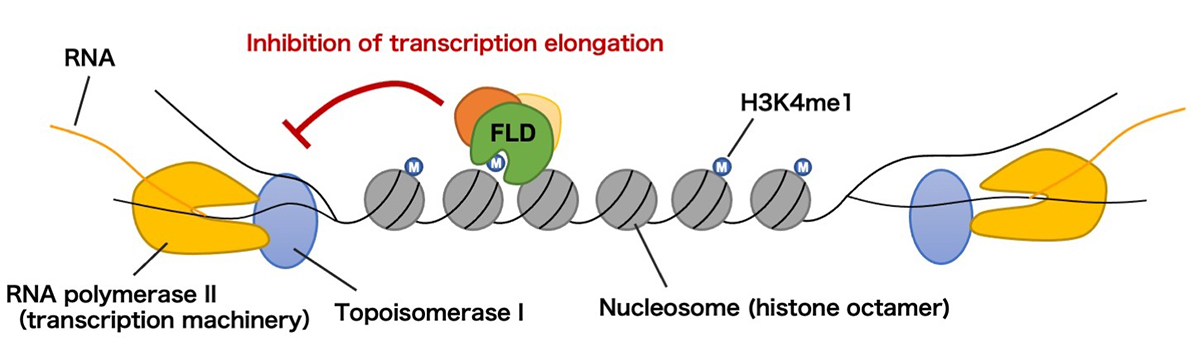
Figure: Proposed model of the findings.
RNA polymerase II transcribes DNA with opening the double helix. Topoisomerase I resolves DNA supercoils to assist transcription elongation. H3K4me1 accumulates in the transcribed regions of genes, but in cases where transcription takes place from both directions, FLD removes H3K4me1 to repress transcription elongation and coordinate bidirectional transcription.















