DNA polymerase ε-dependent modulation of replicative helicase activity at the barrier
DNA polymerase ε-dependent modulation of the pausing property of the CMG helicase at the barrier
Kohji Hizume, Shizuko Endo, Sachiko Muramatsu, Takehiko Kobayashi and Hiroyuki Araki
Genes & Development 32:1315-1320, 2018 DOI:10.1101/gad.317073.118
Chromosome DNA is replicated once per cell cycle. DNA replication starts from many origins spread over chromosomes and replication forks are established. Replication forks often face and pause at the obstacles on chromosomes, such as DNA damage and tightly bound proteins. The proper pausing of replication forks at barriers on chromosomes is important for genome integrity. However, the detailed mechanism underlying this process has not been well elucidated. We successfully reconstituted fork-pausing reactions from purified yeast proteins on templates that had binding sites for the proteins; the forks paused specifically at the protein-bound sites. Moreover, although the replicative CMG helicase alone unwound the protein-bound templates, the unwinding of the protein-bound site was impeded by the presence of a major leading-strand DNA polymerase, Polε. This suggests that Polε modulates CMG to pause at these sites.
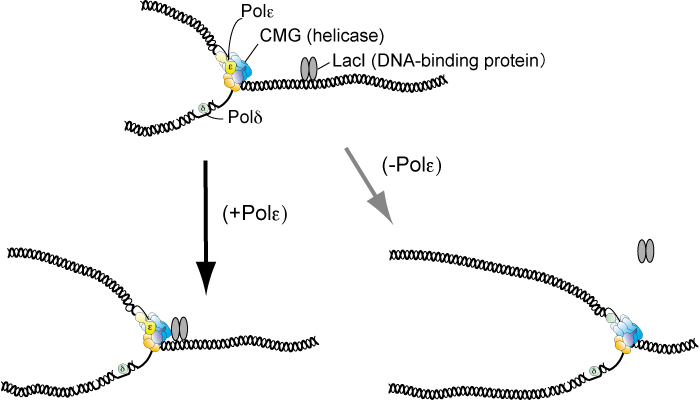
Figure: DNA polymerase ε (Polε) impedes the replicative helicase (CMG) at the site that the protein (LacI) binds and eventually DNA replication stalls. In the absence of Polε, CMG unwinds double-stranded DNA even at the site and thus replication continues.















